An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila
- PMID: 23382693
- PMCID: PMC3561102
- DOI: 10.1371/journal.pgen.1003238
An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila
Abstract
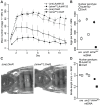
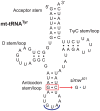
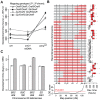
Mitochondrial transcription, translation, and respiration require interactions between genes encoded in two distinct genomes, generating the potential for mutations in nuclear and mitochondrial genomes to interact epistatically and cause incompatibilities that decrease fitness. Mitochondrial-nuclear epistasis for fitness has been documented within and between populations and species of diverse taxa, but rarely has the genetic or mechanistic basis of these mitochondrial-nuclear interactions been elucidated, limiting our understanding of which genes harbor variants causing mitochondrial-nuclear disruption and of the pathways and processes that are impacted by mitochondrial-nuclear coevolution. Here we identify an amino acid polymorphism in the Drosophila melanogaster nuclear-encoded mitochondrial tyrosyl-tRNA synthetase that interacts epistatically with a polymorphism in the D. simulans mitochondrial-encoded tRNA(Tyr) to significantly delay development, compromise bristle formation, and decrease fecundity. The incompatible genotype specifically decreases the activities of oxidative phosphorylation complexes I, III, and IV that contain mitochondrial-encoded subunits. Combined with the identity of the interacting alleles, this pattern indicates that mitochondrial protein translation is affected by this interaction. Our findings suggest that interactions between mitochondrial tRNAs and their nuclear-encoded tRNA synthetases may be targets of compensatory molecular evolution. Human mitochondrial diseases are often genetically complex and variable in penetrance, and the mitochondrial-nuclear interaction we document provides a plausible mechanism to explain this complexity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A Drosophila model for mito-nuclear diseases generated by an incompatible interaction between tRNA and tRNA synthetase.Dis Model Mech. 2015 Aug 1;8(8):843-54. doi: 10.1242/dmm.019323. Epub 2015 May 5. Dis Model Mech. 2015. PMID: 26035388 Free PMC article.
-
Pleiotropic effects of a mitochondrial-nuclear incompatibility depend upon the accelerating effect of temperature in Drosophila.Genetics. 2013 Nov;195(3):1129-39. doi: 10.1534/genetics.113.154914. Epub 2013 Sep 11. Genetics. 2013. PMID: 24026098 Free PMC article.
-
Cytonuclear Interactions in the Evolution of Animal Mitochondrial tRNA Metabolism.Genome Biol Evol. 2015 Jun 27;7(8):2089-101. doi: 10.1093/gbe/evv124. Genome Biol Evol. 2015. PMID: 26116918 Free PMC article.
-
Mitochondrial Aminoacyl-tRNA Synthetase and Disease: The Yeast Contribution for Functional Analysis of Novel Variants.Int J Mol Sci. 2021 Apr 26;22(9):4524. doi: 10.3390/ijms22094524. Int J Mol Sci. 2021. PMID: 33926074 Free PMC article. Review.
-
Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria.Curr Genet. 2009 Feb;55(1):1-18. doi: 10.1007/s00294-008-0223-9. Epub 2008 Dec 16. Curr Genet. 2009. PMID: 19083240 Review.
Cited by
-
Mitonuclear Ecology.Mol Biol Evol. 2015 Aug;32(8):1917-27. doi: 10.1093/molbev/msv104. Epub 2015 Apr 29. Mol Biol Evol. 2015. PMID: 25931514 Free PMC article. Review.
-
Mitonuclear coevolution as the genesis of speciation and the mitochondrial DNA barcode gap.Ecol Evol. 2016 Jul 22;6(16):5831-42. doi: 10.1002/ece3.2338. eCollection 2016 Aug. Ecol Evol. 2016. PMID: 27547358 Free PMC article.
-
The Role of Mitonuclear Incompatibility in Bipolar Disorder Susceptibility and Resilience Against Environmental Stressors.Front Genet. 2021 Mar 16;12:636294. doi: 10.3389/fgene.2021.636294. eCollection 2021. Front Genet. 2021. PMID: 33815470 Free PMC article. Review.
-
Identification of a Novel Variant in EARS2 Associated with a Severe Clinical Phenotype Expands the Clinical Spectrum of LTBL.Genes (Basel). 2020 Sep 2;11(9):1028. doi: 10.3390/genes11091028. Genes (Basel). 2020. PMID: 32887222 Free PMC article.
-
Assessing the fitness consequences of mitonuclear interactions in natural populations.Biol Rev Camb Philos Soc. 2019 Jun;94(3):1089-1104. doi: 10.1111/brv.12493. Epub 2018 Dec 26. Biol Rev Camb Philos Soc. 2019. PMID: 30588726 Free PMC article.
References
-
- Dowling DK, Abiega KC, Arnqvist G (2007) Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution 61: 194–201. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

