Genome-wide haplotype analysis of cis expression quantitative trait loci in monocytes
- PMID: 23382694
- PMCID: PMC3561129
- DOI: 10.1371/journal.pgen.1003240
Genome-wide haplotype analysis of cis expression quantitative trait loci in monocytes
Abstract
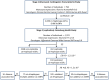
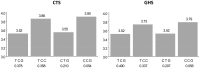
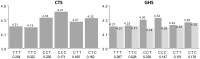
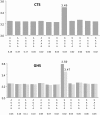
In order to assess whether gene expression variability could be influenced by several SNPs acting in cis, either through additive or more complex haplotype effects, a systematic genome-wide search for cis haplotype expression quantitative trait loci (eQTL) was conducted in a sample of 758 individuals, part of the Cardiogenics Transcriptomic Study, for which genome-wide monocyte expression and GWAS data were available. 19,805 RNA probes were assessed for cis haplotypic regulation through investigation of ~2,1 × 10(9) haplotypic combinations. 2,650 probes demonstrated haplotypic p-values >10(4)-fold smaller than the best single SNP p-value. Replication of significant haplotype effects were tested for 412 probes for which SNPs (or proxies) that defined the detected haplotypes were available in the Gutenberg Health Study composed of 1,374 individuals. At the Bonferroni correction level of 1.2 × 10(-4) (~0.05/412), 193 haplotypic signals replicated. 1000 G imputation was then conducted, and 105 haplotypic signals still remained more informative than imputed SNPs. In-depth analysis of these 105 cis eQTL revealed that at 76 loci genetic associations were compatible with additive effects of several SNPs, while for the 29 remaining regions data could be compatible with a more complex haplotypic pattern. As 24 of the 105 cis eQTL have previously been reported to be disease-associated loci, this work highlights the need for conducting haplotype-based and 1000 G imputed cis eQTL analysis before commencing functional studies at disease-associated loci.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Li J, Burmeister M (2005) Genetical genomics: combining genetics with gene expression analysis. Hum Mol Genet 14 Spec No. 2: R163–169. - PubMed
-
- Montgomery SB, Dermitzakis ET (2009) The resolution of the genetics of gene expression. Hum Mol Genet 18: R211–215. - PubMed
-
- Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, et al. (2007) Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 39: 1208–1216. - PubMed
-
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. (2007) A genome-wide association study of global gene expression. Nat Genet 39: 1202–1207. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

