Small molecule inhibitors of CXCR4
- PMID: 23382786
- PMCID: PMC3563081
- DOI: 10.7150/thno.5376
Small molecule inhibitors of CXCR4
Abstract
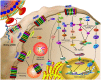
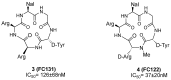
CXCR4 is a G-protein-coupled receptor involved in a number of physiological processes in the hematopoietic and immune systems. The SDF-1/CXCR4 axis is significantly associated with several diseases, such as HIV, cancer, WHIM syndrome, rheumatoid arthritis, pulmonary fibrosis and lupus. For example, CXCR4 is one of the major co-receptors for HIV entry into target cells, while in cancer it plays an important role in tumor cell metastasis. Several promising CXCR4 antagonists have been developed to block SDF-1/CXCR4 interactions that are currently under different stages of development. The first in class CXCR4 antagonist, plerixafor, was approved by the FDA in 2008 for the mobilization of hematopoietic stem cells and several other drugs are currently in clinical trials for cancer, HIV, and WHIM syndrome. While the long-term safety data for the first generation CXCR4 antagonists are not yet available, several new compounds are under preclinical development in an attempt to provide safer and more efficient treatment options for HIV and cancer patients.
Keywords: CXCR4; HIV; WHIM syndrome; antagonists; cancer; lupus.; rheumatoid arthritis.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

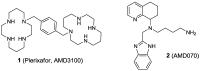
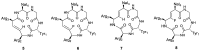




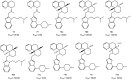
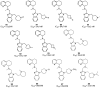

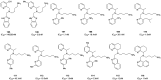





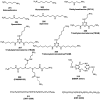
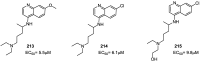

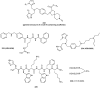
References
-
- Zhan W, Liang Z, Zhu A, Kurtkaya S, Shim H, Snyder JP. et al. Discovery of small molecule CXCR4 antagonists. J Med Chem. 2007;50:5655–64. - PubMed
-
- Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F. et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–5. - PubMed
-
- Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J. et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

