Genetic diversity in the modern horse illustrated from genome-wide SNP data
- PMID: 23383025
- PMCID: PMC3559798
- DOI: 10.1371/journal.pone.0054997
Genetic diversity in the modern horse illustrated from genome-wide SNP data
Abstract
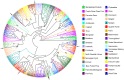
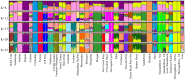
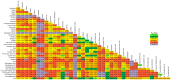
Horses were domesticated from the Eurasian steppes 5,000-6,000 years ago. Since then, the use of horses for transportation, warfare, and agriculture, as well as selection for desired traits and fitness, has resulted in diverse populations distributed across the world, many of which have become or are in the process of becoming formally organized into closed, breeding populations (breeds). This report describes the use of a genome-wide set of autosomal SNPs and 814 horses from 36 breeds to provide the first detailed description of equine breed diversity. F(ST) calculations, parsimony, and distance analysis demonstrated relationships among the breeds that largely reflect geographic origins and known breed histories. Low levels of population divergence were observed between breeds that are relatively early on in the process of breed development, and between those with high levels of within-breed diversity, whether due to large population size, ongoing outcrossing, or large within-breed phenotypic diversity. Populations with low within-breed diversity included those which have experienced population bottlenecks, have been under intense selective pressure, or are closed populations with long breed histories. These results provide new insights into the relationships among and the diversity within breeds of horses. In addition these results will facilitate future genome-wide association studies and investigations into genomic targets of selection.
Conflict of interest statement
Figures

References
-
- FAOSTAT (2010). Available: http://www.faostat.fao.org.Accessed 2012 Jun 5.
-
- Outram AK, Stear NA, Bendrey R, Olsen S, Kasparov A, et al. (2009) The earliest horse harnessing and milking. Science 323: 1332–1335. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

