Modular architecture of metabolic pathways revealed by conserved sequences of reactions
- PMID: 23384306
- PMCID: PMC3632090
- DOI: 10.1021/ci3005379
Modular architecture of metabolic pathways revealed by conserved sequences of reactions
Abstract
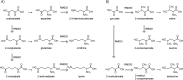
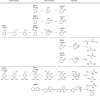
The metabolic network is both a network of chemical reactions and a network of enzymes that catalyze reactions. Toward better understanding of this duality in the evolution of the metabolic network, we developed a method to extract conserved sequences of reactions called reaction modules from the analysis of chemical compound structure transformation patterns in all known metabolic pathways stored in the KEGG PATHWAY database. The extracted reaction modules are repeatedly used as if they are building blocks of the metabolic network and contain chemical logic of organic reactions. Furthermore, the reaction modules often correspond to traditional pathway modules defined as sets of enzymes in the KEGG MODULE database and sometimes to operon-like gene clusters in prokaryotic genomes. We identified well-conserved, possibly ancient, reaction modules involving 2-oxocarboxylic acids. The chain extension module that appears as the tricarboxylic acid (TCA) reaction sequence in the TCA cycle is now shown to be used in other pathways together with different types of modification modules. We also identified reaction modules and their connection patterns for aromatic ring cleavages in microbial biodegradation pathways, which are most characteristic in terms of both distinct reaction sequences and distinct gene clusters. The modular architecture of biodegradation modules will have a potential for predicting degradation pathways of xenobiotic compounds. The collection of these and many other reaction modules is made available as part of the KEGG database.
Figures

Similar articles
-
Chemical and genomic evolution of enzyme-catalyzed reaction networks.FEBS Lett. 2013 Sep 2;587(17):2731-7. doi: 10.1016/j.febslet.2013.06.026. Epub 2013 Jun 28. FEBS Lett. 2013. PMID: 23816707 Review.
-
A new network representation of the metabolism to detect chemical transformation modules.BMC Bioinformatics. 2015 Nov 14;16:385. doi: 10.1186/s12859-015-0809-4. BMC Bioinformatics. 2015. PMID: 26573681 Free PMC article.
-
A systematic comparison of the MetaCyc and KEGG pathway databases.BMC Bioinformatics. 2013 Mar 27;14:112. doi: 10.1186/1471-2105-14-112. BMC Bioinformatics. 2013. PMID: 23530693 Free PMC article.
-
KEGG: integrating viruses and cellular organisms.Nucleic Acids Res. 2021 Jan 8;49(D1):D545-D551. doi: 10.1093/nar/gkaa970. Nucleic Acids Res. 2021. PMID: 33125081 Free PMC article.
-
The compositional and evolutionary logic of metabolism.Phys Biol. 2013 Feb;10(1):011001. doi: 10.1088/1478-3975/10/1/011001. Epub 2012 Dec 12. Phys Biol. 2013. PMID: 23234798 Review.
Cited by
-
Structure Based Virtual Screening Studies to Identify Novel Potential Compounds for GPR142 and Their Relative Dynamic Analysis for Study of Type 2 Diabetes.Front Chem. 2018 Feb 14;6:23. doi: 10.3389/fchem.2018.00023. eCollection 2018. Front Chem. 2018. PMID: 29492402 Free PMC article.
-
Deciphering the Biochemical Pathway and Pharmacokinetic Study of Amyloid βeta-42 with Superparamagnetic Iron Oxide Nanoparticles (SPIONs) Using Systems Biology Approach.Mol Neurobiol. 2018 Apr;55(4):3224-3236. doi: 10.1007/s12035-017-0546-y. Epub 2017 May 6. Mol Neurobiol. 2018. PMID: 28478508
-
Nutrient amendments enrich microbial hydrocarbon degradation metagenomic potential in freshwater coastal wetland microcosm experiments.Appl Environ Microbiol. 2025 Jan 31;91(1):e0197224. doi: 10.1128/aem.01972-24. Epub 2024 Dec 9. Appl Environ Microbiol. 2025. PMID: 39651865 Free PMC article.
-
Data, information, knowledge and principle: back to metabolism in KEGG.Nucleic Acids Res. 2014 Jan;42(Database issue):D199-205. doi: 10.1093/nar/gkt1076. Epub 2013 Nov 7. Nucleic Acids Res. 2014. PMID: 24214961 Free PMC article.
-
Genotypic and Phenotypic Diversity among Human Isolates of Akkermansia muciniphila.mBio. 2021 May 18;12(3):e00478-21. doi: 10.1128/mBio.00478-21. mBio. 2021. PMID: 34006653 Free PMC article.
References
-
- Bono H.; Ogata H.; Goto S.; Kanehisa M. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 1998, 8, 203–210. - PubMed
-
- Galperin M. Y.; Koonin E. V. Functional genomics and enzyme evolution. Homologous and analogous enzymes encoded in microbial genomes. Genetica 1999, 106, 159–170. - PubMed
-
- Forst C. V.; Schulten K. Evolution of metabolisms: a new method for the comparison of metabolic pathways using genomics information. J. Comput. Biol. 1999, 6, 343–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

