Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli
- PMID: 23385483
- PMCID: PMC3588905
- DOI: 10.1038/msb.2012.76
Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli
Abstract
Bacterial populations have a remarkable capacity to cope with extreme environmental fluctuations in their natural environments. In certain cases, adaptation to one stressful environment provides a fitness advantage when cells are exposed to a second stressor, a phenomenon that has been coined as cross-stress protection. A tantalizing question in bacterial physiology is how the cross-stress behavior emerges during evolutionary adaptation and what the genetic basis of acquired stress resistance is. To address these questions, we evolved Escherichia coli cells over 500 generations in five environments that include four abiotic stressors. Through growth profiling and competition assays, we identified several cases of positive and negative cross-stress behavior that span all strain-stress combinations. Resequencing the genomes of the evolved strains resulted in the identification of several mutations and gene amplifications, whose fitness effect was further assessed by mutation reversal and competition assays. Transcriptional profiling of all strains under a specific stress, NaCl-induced osmotic stress, and integration with resequencing data further elucidated the regulatory responses and genes that are involved in this phenomenon. Our results suggest that cross-stress dependencies are ubiquitous, highly interconnected, and can emerge within short timeframes. The high adaptive potential that we observed argues that bacterial populations occupy a genotypic space that enables a high phenotypic plasticity during adaptation in fluctuating environments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

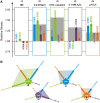


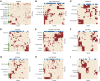
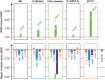
References
-
- Ahuja U, Thony-Meyer L (2005) CcmD is involved in complex formation between CcmC and the heme chaperone CcmE during cytochrome c maturation. J Biol Chem 280: 236–243 - PubMed
-
- Alcántara-Díaz D, Breña-Valle M, Serment-Guerrero J (2004) Divergent adaptation of Escherichia coli to cyclic ultraviolet light exposures. Mutagenesis 19: 349–354 - PubMed
-
- Andrews SC, Robinson AK, Rodriguez-Quinones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237 - PubMed
-
- Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

