Properties of cell death models calibrated and compared using Bayesian approaches
- PMID: 23385484
- PMCID: PMC3588908
- DOI: 10.1038/msb.2012.69
Properties of cell death models calibrated and compared using Bayesian approaches
Abstract
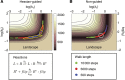
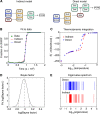
Using models to simulate and analyze biological networks requires principled approaches to parameter estimation and model discrimination. We use Bayesian and Monte Carlo methods to recover the full probability distributions of free parameters (initial protein concentrations and rate constants) for mass-action models of receptor-mediated cell death. The width of the individual parameter distributions is largely determined by non-identifiability but covariation among parameters, even those that are poorly determined, encodes essential information. Knowledge of joint parameter distributions makes it possible to compute the uncertainty of model-based predictions whereas ignoring it (e.g., by treating parameters as a simple list of values and variances) yields nonsensical predictions. Computing the Bayes factor from joint distributions yields the odds ratio (∼20-fold) for competing 'direct' and 'indirect' apoptosis models having different numbers of parameters. Our results illustrate how Bayesian approaches to model calibration and discrimination combined with single-cell data represent a generally useful and rigorous approach to discriminate between competing hypotheses in the face of parametric and topological uncertainty.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Akaike H (1974) A new look at the statistical model identification. Ieee Trans Automat Contr 19: 716–723
-
- Akaike H (1977) On Entropy Maximization Principle Amsterdam: North-Holland,
-
- Akaike H (1983) Information measures and model selection. Bull Int Stat Inst 50: 277–291
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

