The microbiome extends to subepidermal compartments of normal skin
- PMID: 23385576
- PMCID: PMC3655727
- DOI: 10.1038/ncomms2441
The microbiome extends to subepidermal compartments of normal skin
Abstract
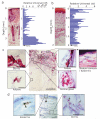
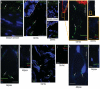
Commensal microbes on the skin surface influence the behaviour of cells below the epidermis. We hypothesized that bacteria or their products exist below the surface epithelium and thus permit physical interaction between microbes and dermal cells. Here to test this hypothesis, we employed multiple independent detection techniques for bacteria including quantitative PCR, Gram staining, immunofluorescence and in situ hybridization. Bacteria were consistently detectable within the dermis and dermal adipose of normal human skin. Sequencing of DNA from dermis and dermal adipose tissue identified bacterial 16S ribosomal RNA reflective of a diverse and partially distinct microbial community in each skin compartment. These results show the microbiota extends within the dermis, therefore, enabling physical contact between bacteria and various cells below the basement membrane. These observations show that normal commensal bacterial communities directly communicate with the host in a tissue previously thought to be sterile.
Figures


References
-
- Wilhelm KP, Cua AB, Maibach HI. Skin aging. Effect on transepidermal water loss, stratum corneum hydration, skin surface pH, and casual sebum content. Arch Dermatol. 1991;127:1806–1809. - PubMed
-
- Ali RS, et al. Expression of the peptide antibiotics human beta defensin-1 and human beta defensin-2 in normal human skin. J Invest Dermatol. 2001;117:106–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

