In Vitro Assessment of the Inflammatory Breast Cancer Cell Line SUM 149: Discovery of 2 Single Nucleotide Polymorphisms in the RNase L Gene
- PMID: 23386909
- PMCID: PMC3563072
- DOI: 10.7150/jca.5002
In Vitro Assessment of the Inflammatory Breast Cancer Cell Line SUM 149: Discovery of 2 Single Nucleotide Polymorphisms in the RNase L Gene
Abstract
Background: Inflammatory breast cancer (IBC) is a rare, highly aggressive form of breast cancer. The mechanism of IBC carcinogenesis remains unknown. We sought to evaluate potential genetic risk factors for IBC and whether or not the IBC cell lines SUM149 and SUM190 demonstrated evidence of viral infection.
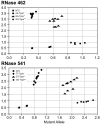
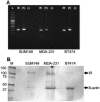
Methods: We performed single nucleotide polymorphism (SNP) genotyping for 2 variants of the ribonuclease (RNase) L gene that have been correlated with the risk of prostate cancer due to a possible viral etiology. We evaluated dose-response to treatment with interferon-alpha (IFN-α); and assayed for evidence of the putative human mammary tumor virus (HMTV, which has been implicated in IBC) in SUM149 cells. A bioinformatic analysis was performed to evaluate expression of RNase L in IBC and non-IBC.
Results: 2 of 2 IBC cell lines were homozygous for RNase L common missense variants 462 and 541; whereas 2 of 10 non-IBC cell lines were homozygous positive for the 462 variant (p= 0.09) and 0 of 10 non-IBC cell lines were homozygous positive for the 541 variant (p = 0.015). Our real-time polymerase chain reaction (RT-PCR) and Southern blot analysis for sequences of HMTV revealed no evidence of the putative viral genome.
Conclusion: We discovered 2 SNPs in the RNase L gene that were homozygously present in IBC cell lines. The 462 variant was absent in non-IBC lines. Our discovery of these SNPs present in IBC cell lines suggests a possible biomarker for risk of IBC. We found no evidence of HMTV in SUM149 cells. A query of a panel of human IBC and non-IBC samples showed no difference in RNase L expression. Further studies of the RNase L 462 and 541 variants in IBC tissues are warranted to validate our in vitro findings.
Keywords: HMTV; MMTV; RNase L.; SUM149; inflammatory breast cancer; interferon-alpha.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Gonzalez-Angulo AM, Hennessy BT, Broglio K. et al. Trends for inflammatory breast cancer: is survival improving? Oncologist. 2007;12:904–12. - PubMed
-
- Charafe-Jauffret E, Tarpin C, Viens P, Bertucci F. Defining the molecular biology of inflammatory breast cancer. Semin Oncol. 2008;35:41–50. - PubMed
-
- Dirix LY, Van Dam P, Prove A, Vermeulen PB. Inflammatory breast cancer: current understanding. Curr Opin Oncol. 2006;18:563–71. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

