Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities
- PMID: 23387867
- PMCID: PMC3665634
- DOI: 10.1111/1462-2920.12086
Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities
Abstract
Next-generation sequencing has dramatically changed the landscape of microbial ecology, large-scale and in-depth diversity studies being now widely accessible. However, determining the accuracy of taxonomic and quantitative inferences and comparing results obtained with different approaches are complicated by incongruence of experimental and computational data types and also by lack of knowledge of the true ecological diversity. Here we used highly diverse bacterial and archaeal synthetic communities assembled from pure genomic DNAs to compare inferences from metagenomic and SSU rRNA amplicon sequencing. Both Illumina and 454 metagenomic data outperformed amplicon sequencing in quantifying the community composition, but the outcome was dependent on analysis parameters and platform. New approaches in processing and classifying amplicons can reconstruct the taxonomic composition of the community with high reproducibility within primer sets, but all tested primers sets lead to significant taxon-specific biases. Controlled synthetic communities assembled to broadly mimic the phylogenetic richness in target environments can provide important validation for fine-tuning experimental and computational parameters used to characterize natural communities.
© 2013 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures
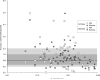
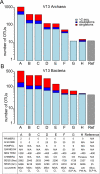

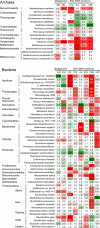
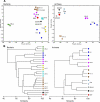
References
-
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

