Impact of deleterious passenger mutations on cancer progression
- PMID: 23388632
- PMCID: PMC3581883
- DOI: 10.1073/pnas.1213968110
Impact of deleterious passenger mutations on cancer progression
Abstract
Cancer progression is driven by the accumulation of a small number of genetic alterations. However, these few driver alterations reside in a cancer genome alongside tens of thousands of additional mutations termed passengers. Passengers are widely believed to have no role in cancer, yet many passengers fall within protein-coding genes and other functional elements that can have potentially deleterious effects on cancer cells. Here we investigate the potential of moderately deleterious passengers to accumulate and alter the course of neoplastic progression. Our approach combines evolutionary simulations of cancer progression with an analysis of cancer sequencing data. From simulations, we find that passengers accumulate and largely evade natural selection during progression. Although individually weak, the collective burden of passengers alters the course of progression, leading to several oncological phenomena that are hard to explain with a traditional driver-centric view. We then tested the predictions of our model using cancer genomics data and confirmed that many passengers are likely damaging and have largely evaded negative selection. Finally, we use our model to explore cancer treatments that exploit the load of passengers by either (i) increasing the mutation rate or (ii) exacerbating their deleterious effects. Though both approaches lead to cancer regression, the latter is a more effective therapy. Our results suggest a unique framework for understanding cancer progression as a balance of driver and passenger mutations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

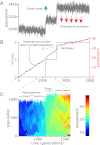
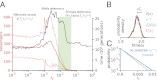
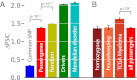
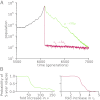
Comment in
-
Tumour evolution: Weighed down by passengers?Nat Rev Cancer. 2013 Apr;13(4):219. doi: 10.1038/nrc3488. Epub 2013 Mar 7. Nat Rev Cancer. 2013. PMID: 23467303 No abstract available.
References
-
- Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

