Critical residues in the PMEL/Pmel17 N-terminus direct the hierarchical assembly of melanosomal fibrils
- PMID: 23389629
- PMCID: PMC3608505
- DOI: 10.1091/mbc.E12-10-0742
Critical residues in the PMEL/Pmel17 N-terminus direct the hierarchical assembly of melanosomal fibrils
Abstract
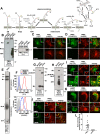
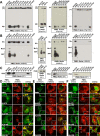
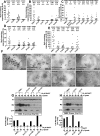
PMEL (also called Pmel17 or gp100) is a melanocyte/melanoma-specific glycoprotein that plays a critical role in melanosome development by forming a fibrillar amyloid matrix in the organelle for melanin deposition. Although ultimately not a component of mature fibrils, the PMEL N-terminal region (NTR) is essential for their formation. By mutational analysis we establish a high-resolution map of this domain in which sequence elements and functionally critical residues are assigned. We show that the NTR functions in cis to drive the aggregation of the downstream polycystic kidney disease (PKD) domain into a melanosomal core matrix. This is essential to promote in trans the stabilization and terminal proteolytic maturation of the repeat (RPT) domain-containing MαC units, precursors of the second fibrillogenic fragment. We conclude that during melanosome biogenesis the NTR controls the hierarchical assembly of melanosomal fibrils.
Figures






Similar articles
-
Lysophospholipid-containing membranes modulate the fibril formation of the repeat domain of a human functional amyloid, pmel17.J Mol Biol. 2014 Dec 12;426(24):4074-4086. doi: 10.1016/j.jmb.2014.10.009. Epub 2014 Oct 14. J Mol Biol. 2014. PMID: 25451784 Free PMC article.
-
Biogenesis of fibrils requires C-mannosylation of PMEL.FEBS J. 2023 Nov;290(22):5373-5394. doi: 10.1111/febs.16927. Epub 2023 Aug 18. FEBS J. 2023. PMID: 37552474
-
PMEL Amyloid Fibril Formation: The Bright Steps of Pigmentation.Int J Mol Sci. 2016 Aug 31;17(9):1438. doi: 10.3390/ijms17091438. Int J Mol Sci. 2016. PMID: 27589732 Free PMC article. Review.
-
The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers.J Biol Chem. 2006 Jul 28;281(30):21198-21208. doi: 10.1074/jbc.M601643200. Epub 2006 May 8. J Biol Chem. 2006. PMID: 16682408
-
Why Study Functional Amyloids? Lessons from the Repeat Domain of Pmel17.J Mol Biol. 2018 Oct 12;430(20):3696-3706. doi: 10.1016/j.jmb.2018.06.011. Epub 2018 Jun 7. J Mol Biol. 2018. PMID: 29886018 Free PMC article. Review.
Cited by
-
The PKD domain distinguishes the trafficking and amyloidogenic properties of the pigment cell protein PMEL and its homologue GPNMB.Pigment Cell Melanoma Res. 2013 Jul;26(4):470-86. doi: 10.1111/pcmr.12084. Epub 2013 Apr 2. Pigment Cell Melanoma Res. 2013. PMID: 23452376 Free PMC article.
-
Three tapasin docking sites in TAP cooperate to facilitate transporter stabilization and heterodimerization.J Immunol. 2014 Mar 1;192(5):2480-94. doi: 10.4049/jimmunol.1302637. Epub 2014 Feb 5. J Immunol. 2014. PMID: 24501197 Free PMC article.
-
Amyloidosis in alkaptonuria.J Inherit Metab Dis. 2015 Sep;38(5):797-805. doi: 10.1007/s10545-015-9842-8. Epub 2015 Apr 14. J Inherit Metab Dis. 2015. PMID: 25868666 Review.
-
Cryo-EM of wild-type and mutant PMEL amyloid cores reveals structural mechanism of pigment dispersion syndrome.Nat Commun. 2025 Jul 1;16(1):5411. doi: 10.1038/s41467-025-61233-y. Nat Commun. 2025. PMID: 40595665 Free PMC article.
-
Molecular origin of pH-dependent fibril formation of a functional amyloid.Chembiochem. 2014 Jul 21;15(11):1569-72. doi: 10.1002/cbic.201402074. Epub 2014 Jun 20. Chembiochem. 2014. PMID: 24954152 Free PMC article.
References
-
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

