Burkholderia cenocepacia conditional growth mutant library created by random promoter replacement of essential genes
- PMID: 23389959
- PMCID: PMC3633349
- DOI: 10.1002/mbo3.71
Burkholderia cenocepacia conditional growth mutant library created by random promoter replacement of essential genes
Abstract
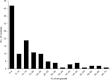
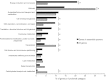
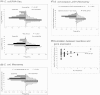
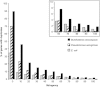
Identification of essential genes by construction of conditional knockouts with inducible promoters allows the identification of essential genes and creation of conditional growth (CG) mutants that are then available as genetic tools for further studies. We used large-scale transposon delivery of the rhamnose-inducible promoter, PrhaB followed by robotic screening of rhamnose-dependent growth to construct a genomic library of 106 Burkholderia cenocepacia CG mutants. Transposon insertions were found where PrhaB was in the same orientation of widely conserved, well-characterized essential genes as well as genes with no previous records of essentiality in other microorganisms. Using previously reported global gene-expression analyses, we demonstrate that PrhaB can achieve the wide dynamic range of expression levels required for essential genes when the promoter is delivered randomly and mutants with rhamnose-dependent growth are selected. We also show specific detection of the target of an antibiotic, novobiocin, by enhanced sensitivity of the corresponding CG mutant (PrhaB controlling gyrB expression) within the library. Modulation of gene expression to achieve 30-60% of wild-type growth created conditions for specific hypersensitivity demonstrating the value of the CG mutant library for chemogenomic experiments. In summary, CG mutants can be obtained on a large scale by random delivery of a tightly regulated inducible promoter into the bacterial chromosome followed by a simple screening for the CG phenotype, without previous information on gene essentiality.
© 2013 The Authors. Published by Blackwell Publishing Ltd.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Baba T, Huan HC, Datsenko K, Wanner BL, Mori H. The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Methods Mol. Biol. 2008;416:183–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

