Time line of redox events in aging postmitotic cells
- PMID: 23390587
- PMCID: PMC3564446
- DOI: 10.7554/eLife.00306
Time line of redox events in aging postmitotic cells
Abstract
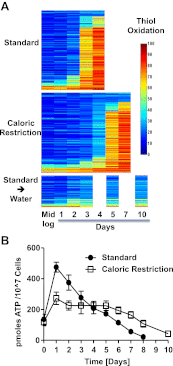
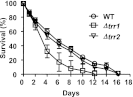
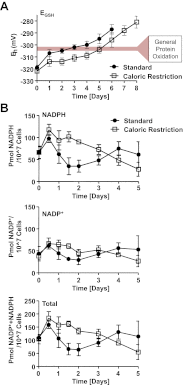
The precise roles that oxidants play in lifespan and aging are still unknown. Here, we report the discovery that chronologically aging yeast cells undergo a sudden redox collapse, which affects over 80% of identified thiol-containing proteins. We present evidence that this redox collapse is not triggered by an increase in endogenous oxidants as would have been postulated by the free radical theory of aging. Instead it appears to be instigated by a substantial drop in cellular NADPH, which normally provides the electron source for maintaining cellular redox homeostasis. This decrease in NADPH levels occurs very early during lifespan and sets into motion a cascade that is predicted to down-regulate most cellular processes. Caloric restriction, a near-universal lifespan extending measure, increases NADPH levels and delays each facet of the cascade. Our studies reveal a time line of events leading up to the system-wide oxidation of the proteome days before cell death.DOI:http://dx.doi.org/10.7554/eLife.00306.001.
Keywords: Aging; Metabolism; Oxidative Stress; Redox Proteomics; Redox Regulation; S. cerevisiae.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Comment in
- doi: 10.7554/eLife.00515
References
-
- Barja G. 2002. Endogenous oxidative stress: relationship to aging, longevity and caloric restriction. Ageing Res Rev 1:397–411 - PubMed
-
- Bondar RJ, Mead DC. 1974. Evaluation of glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides in the hexokinase method for determining glucose in serum. Clin Chem 20:586–90 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

