Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification
- PMID: 23391577
- PMCID: PMC3613443
- DOI: 10.1104/pp.112.208694
Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification
Abstract
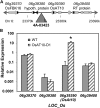
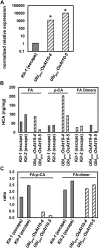
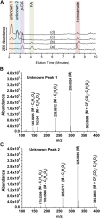
Grass cell wall properties influence food, feed, and biofuel feedstock usage efficiency. The glucuronoarabinoxylan of grass cell walls is esterified with the phenylpropanoid-derived hydroxycinnamic acids ferulic acid (FA) and para-coumaric acid (p-CA). Feruloyl esters undergo oxidative coupling with neighboring phenylpropanoids on glucuronoarabinoxylan and lignin. Examination of rice (Oryza sativa) mutants in a grass-expanded and -diverged clade of BAHD acyl-coenzyme A-utilizing transferases identified four mutants with altered cell wall FA or p-CA contents. Here, we report on the effects of overexpressing one of these genes, OsAt10 (LOC_Os06g39390), in rice. An activation-tagged line, OsAT10-D1, shows a 60% reduction in matrix polysaccharide-bound FA and an approximately 300% increase in p-CA in young leaf tissue but no discernible phenotypic alterations in vegetative development, lignin content, or lignin composition. Two additional independent OsAt10 overexpression lines show similar changes in FA and p-CA content. Cell wall fractionation and liquid chromatography-mass spectrometry experiments isolate the cell wall alterations in the mutant to ester conjugates of a five-carbon sugar with p-CA and FA. These results suggest that OsAT10 is a p-coumaroyl coenzyme A transferase involved in glucuronoarabinoxylan modification. Biomass from OsAT10-D1 exhibits a 20% to 40% increase in saccharification yield depending on the assay. Thus, OsAt10 is an attractive target for improving grass cell wall quality for fuel and animal feed.
Figures








References
-
- Agblevor FA, Evans RJ, Johnson KD. (1994) Molecular-beam mass-spectrometric analysis of lignocellulosic materials. I. Herbaceous biomass. J Anal Appl Pyrolysis 30: 125–144
-
- Allerdings E, Ralph J, Schatz PF, Gniechwitz D, Steinhart H, Bunzel M. (2005) Isolation and structural identification of diarabinosyl 8-O-4-dehydrodiferulate from maize bran insoluble fibre. Phytochemistry 66: 113–124 - PubMed
-
- An G, Lee S, Kim SH, Kim SR. (2005) Molecular genetics using T-DNA in rice. Plant Cell Physiol 46: 14–22 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

