PAR-1 contributes to the innate immune response during viral infection
- PMID: 23391721
- PMCID: PMC3582138
- DOI: 10.1172/JCI66125
PAR-1 contributes to the innate immune response during viral infection
Abstract
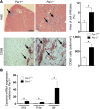
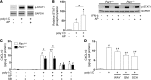
Coagulation is a host defense system that limits the spread of pathogens. Coagulation proteases, such as thrombin, also activate cells by cleaving PARs. In this study, we analyzed the role of PAR-1 in coxsackievirus B3-induced (CVB3-induced) myocarditis and influenza A infection. CVB3-infected Par1(-/-) mice expressed reduced levels of IFN-β and CXCL10 during the early phase of infection compared with Par1(+/+) mice that resulted in higher viral loads and cardiac injury at day 8 after infection. Inhibition of either tissue factor or thrombin in WT mice also significantly increased CVB3 levels in the heart and cardiac injury compared with controls. BM transplantation experiments demonstrated that PAR-1 in nonhematopoietic cells protected mice from CVB3 infection. Transgenic mice overexpressing PAR-1 in cardiomyocytes had reduced CVB3-induced myocarditis. We found that cooperative signaling between PAR-1 and TLR3 in mouse cardiac fibroblasts enhanced activation of p38 and induction of IFN-β and CXCL10 expression. Par1(-/-) mice also had decreased CXCL10 expression and increased viral levels in the lung after influenza A infection compared with Par1(+/+) mice. Our results indicate that the tissue factor/thrombin/PAR-1 pathway enhances IFN-β expression and contributes to the innate immune response during single-stranded RNA viral infection.
Figures


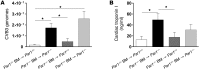

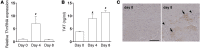



References
-
- Iwanaga S, Kawabata S. Evolution and phylogeny of defense molecules associated with innate immunity in horseshoe crab. Front Biosci. 1998;3:D973–D984. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

