STAT2 deficiency and susceptibility to viral illness in humans
- PMID: 23391734
- PMCID: PMC3581986
- DOI: 10.1073/pnas.1220098110
STAT2 deficiency and susceptibility to viral illness in humans
Abstract
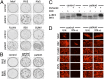
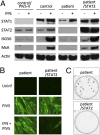
Severe infectious disease in children may be a manifestation of primary immunodeficiency. These genetic disorders represent important experiments of nature with the capacity to elucidate nonredundant mechanisms of human immunity. We hypothesized that a primary defect of innate antiviral immunity was responsible for unusually severe viral illness in two siblings; the proband developed disseminated vaccine strain measles following routine immunization, whereas an infant brother died after a 2-d febrile illness from an unknown viral infection. Patient fibroblasts were indeed abnormally permissive for viral replication in vitro, associated with profound failure of type I IFN signaling and absence of STAT2 protein. Sequencing of genomic DNA and RNA revealed a homozygous mutation in intron 4 of STAT2 that prevented correct splicing in patient cells. Subsequently, other family members were identified with the same genetic lesion. Despite documented infection by known viral pathogens, some of which have been more severe than normal, surviving STAT2-deficient individuals have remained generally healthy, with no obvious defects in their adaptive immunity or developmental abnormalities. These findings imply that type I IFN signaling [through interferon-stimulated gene factor 3 (ISGF3)] is surprisingly not essential for host defense against the majority of common childhood viral infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Randall RE, Goodbourn S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(pt 1):1–47. - PubMed
-
- Müller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. - PubMed
-
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84(3):443–450. - PubMed
-
- Meraz MA, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84(3):431–442. - PubMed
-
- Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13(6):795–804. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

