RNA pseudouridylation: new insights into an old modification
- PMID: 23391857
- PMCID: PMC3608706
- DOI: 10.1016/j.tibs.2013.01.002
RNA pseudouridylation: new insights into an old modification
Abstract
Pseudouridine is the most abundant post-transcriptionally modified nucleotide in various stable RNAs of all organisms. Pseudouridine is derived from uridine via base-specific isomerization, resulting in an extra hydrogen-bond donor that distinguishes it from other nucleotides. In eukaryotes, uridine-to-pseudouridine isomerization is catalyzed primarily by box H/ACA RNPs, ribonucleoproteins that act as pseudouridylases. When introduced into RNA, pseudouridine contributes significantly to RNA-mediated cellular processes. It was recently discovered that pseudouridylation can be induced by stress, suggesting a regulatory role for pseudouridine. It has also been reported that pseudouridine can be artificially introduced into mRNA by box H/ACA RNPs and that such introduction can mediate nonsense-to-sense codon conversion, thus demonstrating a new means of generating coding or protein diversity.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
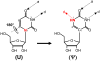






References
-
- Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas: Landes Bioscience; 2009.
-
- Cohn WE, Volkin W. Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature (London) 1951;167:483–484.
-
- Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227(2):907–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

