HMGA1 directly interacts with TAR to modulate basal and Tat-dependent HIV transcription
- PMID: 23392246
- PMCID: PMC3672287
- DOI: 10.4161/rna.23686
HMGA1 directly interacts with TAR to modulate basal and Tat-dependent HIV transcription
Abstract
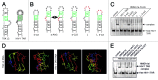
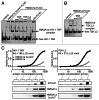
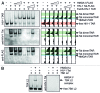
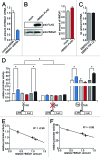
The transactivating response element (TAR) of human immunodeficiency virus 1 (HIV-1) is essential for promoter transactivation by the viral transactivator of transcription (Tat). The Tat-TAR interaction thereby recruits active positive transcription elongation factor b (P-TEFb) from its inactive, 7SK/HEXIM1-bound form, leading to efficient viral transcription. Here, we show that the 7SK RNA-associating chromatin regulator HMGA1 can specifically bind to the HIV-1 TAR element and that 7SK RNA can thereby compete with TAR. The HMGA1-binding interface of TAR is located within the binding site for Tat and other cellular activators, and we further provide evidence for competition between HMGA1 and Tat for TAR-binding. HMGA1 negatively influences the expression of a HIV-1 promoter-driven reporter in a TAR-dependent manner, both in the presence and in the absence of Tat. The overexpression of the HMGA1-binding substructure of 7SK RNA results in a TAR-dependent gain of HIV-1 promoter activity similar to the effect of the shRNA-mediated knockdown of HMGA1. Our results support a model in which the HMGA1/TAR interaction prevents the binding of transcription-activating cellular co-factors and Tat, subsequently leading to reduced HIV-1 transcription.
Keywords: 7SK snRNA; HIV-1; HMGA1; P-TEFb; TAR; Tat; elongation; transcription.
Figures
Similar articles
-
Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat.PLoS Pathog. 2010 Oct 14;6(10):e1001152. doi: 10.1371/journal.ppat.1001152. PLoS Pathog. 2010. PMID: 20976203 Free PMC article.
-
Latent HIV-1 TAR Regulates 7SK-responsive P-TEFb Target Genes and Targets Cellular Immune Responses in the Absence of Tat.Genomics Proteomics Bioinformatics. 2017 Oct;15(5):313-323. doi: 10.1016/j.gpb.2017.05.003. Epub 2017 Oct 14. Genomics Proteomics Bioinformatics. 2017. PMID: 29037489 Free PMC article.
-
HMGA1-dependent and independent 7SK RNA gene regulatory activity.RNA Biol. 2011 Jan-Feb;8(1):143-57. doi: 10.4161/rna.8.1.14261. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21282977
-
New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC).J Neuroimmune Pharmacol. 2011 Jun;6(2):260-8. doi: 10.1007/s11481-011-9267-6. Epub 2011 Mar 1. J Neuroimmune Pharmacol. 2011. PMID: 21360054 Free PMC article. Review.
-
Inhibitors of HIV-1 Tat-mediated transactivation.Curr Med Chem. 2006;13(11):1305-15. doi: 10.2174/092986706776872989. Curr Med Chem. 2006. PMID: 16712471 Review.
Cited by
-
GAS5‑mediated regulation of cell signaling (Review).Mol Med Rep. 2020 Oct;22(4):3049-3056. doi: 10.3892/mmr.2020.11435. Epub 2020 Aug 19. Mol Med Rep. 2020. PMID: 32945519 Free PMC article. Review.
-
Phosphorylation orchestrates the structural ensemble of the intrinsically disordered protein HMGA1a and modulates its DNA binding to the NFκB promoter.Nucleic Acids Res. 2019 Dec 16;47(22):11906-11920. doi: 10.1093/nar/gkz614. Nucleic Acids Res. 2019. PMID: 31340016 Free PMC article.
-
Anti-Transcription Factor RNA Aptamers as Potential Therapeutics.Nucleic Acid Ther. 2016 Feb;26(1):29-43. doi: 10.1089/nat.2015.0566. Epub 2015 Oct 28. Nucleic Acid Ther. 2016. PMID: 26509637 Free PMC article. Review.
-
The new (dis)order in RNA regulation.Cell Commun Signal. 2016 Apr 6;14:9. doi: 10.1186/s12964-016-0132-3. Cell Commun Signal. 2016. PMID: 27048167 Free PMC article. Review.
-
RNA-Mediated Regulation of HMGA1 Function.Biomolecules. 2015;5(2):943-57. doi: 10.3390/biom5020943. Biomolecules. 2015. PMID: 26117853 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
