Novel higher-order epigenetic regulation of the Bdnf gene upon seizures
- PMID: 23392678
- PMCID: PMC6619147
- DOI: 10.1523/JNEUROSCI.1085-12.2013
Novel higher-order epigenetic regulation of the Bdnf gene upon seizures
Erratum in
-
Correction: Walczak et al., "Novel Higher-Order Epigenetic Regulation of the Bdnf Gene upon Seizures".J Neurosci. 2017 Mar 15;37(11):3102. doi: 10.1523/JNEUROSCI.0477-17.2017. J Neurosci. 2017. PMID: 28298573 Free PMC article. No abstract available.
Abstract
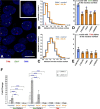
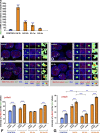
Studies in cultured cells have demonstrated the existence of higher-order epigenetic mechanisms, determining the relationship between expression of the gene and its position within the cell nucleus. It is unknown, whether such mechanisms operate in postmitotic, highly differentiated cell types, such as neurons in vivo. Accordingly, we examined whether the intranuclear positions of Bdnf and Trkb genes, encoding the major neurotrophin and its receptor respectively, change as a result of neuronal activity, and what functional consequences such movements may have. In a rat model of massive neuronal activation upon kainate-induced seizures we found that elevated neuronal expression of Bdnf is associated with its detachment from the nuclear lamina, and translocation toward the nucleus center. In contrast, the position of stably expressed Trkb remains unchanged after seizures. Our study demonstrates that activation-dependent architectural remodeling of the neuronal cell nucleus in vivo contributes to activity-dependent changes in gene expression in the brain.
Figures
References
-
- Barco A, Marie H. Genetic approaches to investigate the role of CREB in neuronal plasticity and memory. Mol Neurobiol. 2011;44:330–349. - PubMed
-
- Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, Lesch KP, Lanfumey L, Steinbusch HW, Kenis G. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2012;17:584–596. - PubMed
-
- Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76:99–125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
