Chromosome evolution in marginal populations of Aegilops speltoides: causes and consequences
- PMID: 23393097
- PMCID: PMC3605956
- DOI: 10.1093/aob/mct023
Chromosome evolution in marginal populations of Aegilops speltoides: causes and consequences
Abstract
Background: Genome restructuring is an ongoing process in natural plant populations. The influence of environmental changes on the genome is crucial, especially during periods of extreme climatic fluctuations. Interactions between the environment and the organism manifest to the greatest extent at the limits of the species' ecological niche. Thus, marginal populations are expected to exhibit lower genetic diversity and higher genetic differentiation than central populations, and some models assume that marginal populations play an important role in the maintenance and generation of biological diversity.
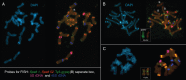
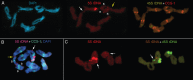
Scope: In this review, long-term data on the cytogenetic characteristics of diploid Aegilops speltoides Tauch populations are summarized and discussed. This species is distributed in and around the Fertile Crescent and is proposed to be the wild progenitor of a number of diploid and polyploid wheat species. In marginal populations of Ae. speltoides, numerical chromosomal aberrations, spontaneous aneuploidy, B-chromosomes, rDNA cluster repatterning and reduction in the species-specific and tribe-specific tandem repeats have been detected. Significant changes were observed and occurred in parallel with changes in plant morphology and physiology.
Conclusions: Considerable genomic variation at the chromosomal level was found in the marginal populations of Ae. speltoides. It is likely that a specific combination of gene mutations and chromosomal repatterning has produced the evolutionary trend in each specific case, i.e. for a particular species or group of related species in a given period of time and in a certain habitat. The appearance of a new chromosomal pattern is considered an important factor in promoting the emergence of interbreeding barriers.
Figures


References
-
- Anamthawat-Jonsson K, Heslop-Harrison JS. Isolation and characterization of genome-specific DNA sequences in Triticeae species. Molecular and General Genetics. 1993;240:151–158. - PubMed
-
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromere sequence. Chromosoma. 1996;105:261–268. - PubMed
-
- Badaeva ED, Amosova AV, Muravenko OV, et al. Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Systematics and Evolution. 2002;231:163–190.
-
- Badaeva ED, Amosova AV, Samatadze TE, et al. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Systematics and Evolution. 2004;246:45–76.
-
- Badaeva ED, Dedkova OS, Gay G, et al. Chromosomal rearrangements in wheat: their types and distribution. Genome. 2007;50:907–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

