Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase
- PMID: 23393192
- PMCID: PMC3616717
- DOI: 10.1093/nar/gkt056
Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase
Abstract
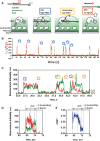
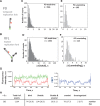
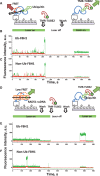
DNA repair helicases function in the cell to separate DNA duplexes or remodel nucleoprotein complexes. These functions are influenced by sensing and signaling; the cellular pool of a DNA helicase may contain subpopulations of enzymes carrying different post-translational modifications and performing distinct biochemical functions. Here, we report a novel experimental strategy, single-molecule sorting, which overcomes difficulties associated with comprehensive analysis of heterologously modified pool of proteins. This methodology was applied to visualize human DNA helicase F-box-containing DNA helicase (FBH1) acting on the DNA structures resembling a stalled or collapsed replication fork and its interactions with RAD51 nucleoprotein filament. Individual helicase molecules isolated from human cells with their native post-translational modifications were analyzed using total internal reflection fluorescence microscopy. Separation of the activity trajectories originated from ubiquitylated and non-ubiquitylated FBH1 molecules revealed that ubiquitylation affects FBH1 interaction with the RAD51 nucleoprotein filament, but not its translocase and helicase activities.
Figures


References
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed. Engl. 2005;44:7342–7372. - PubMed
-
- Thomson TM, Guerra-Rebollo M. Ubiquitin and SUMO signalling in DNA repair. Biochem. Soc. Trans. 2010;38:116–131. - PubMed
-
- van Kasteren S. Synthesis of post-translationally modified proteins. Biochem. Soc. Trans. 2012;40:929–944. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

