Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins
- PMID: 23393263
- PMCID: PMC3899395
- DOI: 10.1126/science.1229934
Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins
Abstract
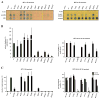
E6 viral oncoproteins are key players in epithelial tumors induced by papillomaviruses in vertebrates, including cervical cancer in humans. E6 proteins target many host proteins by specifically interacting with acidic LxxLL motifs. We solved the crystal structures of bovine (BPV1) and human (HPV16) papillomavirus E6 proteins bound to LxxLL peptides from the focal adhesion protein paxillin and the ubiquitin ligase E6AP, respectively. In both E6 proteins, two zinc domains and a linker helix form a basic-hydrophobic pocket, which captures helical LxxLL motifs in a way compatible with other interaction modes. Mutational inactivation of the LxxLL binding pocket disrupts the oncogenic activities of both E6 proteins. This work reveals the structural basis of both the multifunctionality and the oncogenicity of E6 proteins.
Figures
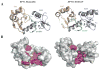

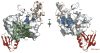
References
-
- zur Hausen H. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin Cancer Biol. 1999;9:405. - PubMed
-
- Pfister H, et al. High prevalence of epidermodysplasia verruciformis-associated human papillomavirus DNA in actinic keratoses of the immunocompetent population. Arch Dermatol Res. 2003;295:273. - PubMed
-
- Nasir L, Campo MS. Bovine papillomaviruses: their role in the aetiology of cutaneous tumours of bovids and equids. Vet Dermatol. 2008;19:243. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

