A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility
- PMID: 23394947
- PMCID: PMC3652613
- DOI: 10.1016/j.cell.2013.01.030
A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility
Abstract
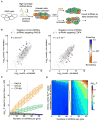
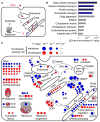
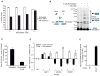
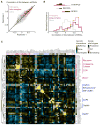
Genetic interaction (GI) maps, comprising pairwise measures of how strongly the function of one gene depends on the presence of a second, have enabled the systematic exploration of gene function in microorganisms. Here, we present a two-stage strategy to construct high-density GI maps in mammalian cells. First, we use ultracomplex pooled shRNA libraries (25 shRNAs/gene) to identify high-confidence hit genes for a given phenotype and effective shRNAs. We then construct double-shRNA libraries from these to systematically measure GIs between hits. A GI map focused on ricin susceptibility broadly recapitulates known pathways and provides many unexpected insights. These include a noncanonical role for COPI, a previously uncharacterized protein complex affecting toxin clearance, a specialized role for the ribosomal protein RPS25, and functionally distinct mammalian TRAPP complexes. The ability to rapidly generate mammalian GI maps provides a potentially transformative tool for defining gene function and designing combination therapies based on synergistic pairs.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Genetic screening: Interaction maps for mammals.Nat Rev Genet. 2013 Apr;14(4):240. doi: 10.1038/nrg3451. Epub 2013 Feb 19. Nat Rev Genet. 2013. PMID: 23419280 No abstract available.
-
Genetics: Mammalian genes interacting.Nat Methods. 2013 Apr;10(4):281. doi: 10.1038/nmeth.2424. Nat Methods. 2013. PMID: 23653921 No abstract available.

