Protein determinants of meiotic DNA break hot spots
- PMID: 23395004
- PMCID: PMC3595357
- DOI: 10.1016/j.molcel.2013.01.008
Protein determinants of meiotic DNA break hot spots
Abstract
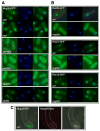
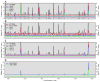
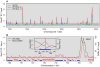
Meiotic recombination, crucial for proper chromosome segregation and genome evolution, is initiated by programmed DNA double-strand breaks (DSBs) in yeasts and likely all sexually reproducing species. In fission yeast, DSBs occur up to hundreds of times more frequently at special sites, called hot spots, than in other regions of the genome. What distinguishes hot spots from cold regions is an unsolved problem, although transcription factors determine some hot spots. We report the discovery that three coiled-coil proteins-Rec25, Rec27, and Mug20-bind essentially all hot spots with great specificity even without DSB formation. These small proteins are components of linear elements, are related to synaptonemal complex proteins, and are essential for nearly all DSBs at most hot spots. Our results indicate these hot spot determinants activate or stabilize the DSB-forming protein Rec12 (Spo11 homolog) rather than promote its binding to hot spots. We propose a paradigm for hot spot determination and crossover control by linear element proteins.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


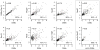
Similar articles
-
Mug20-Rec25-Rec27 binds DNA and enhances meiotic DNA break formation via phase-separated condensates.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf123. doi: 10.1093/nar/gkaf123. Nucleic Acids Res. 2025. PMID: 40037704 Free PMC article.
-
Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination.Curr Biol. 2008 Jun 3;18(11):849-54. doi: 10.1016/j.cub.2008.05.025. Curr Biol. 2008. PMID: 18514516 Free PMC article.
-
Activation of meiotic recombination by nuclear import of the DNA break hotspot-determining complex in fission yeast.J Cell Sci. 2021 Feb 22;134(4):jcs253518. doi: 10.1242/jcs.253518. J Cell Sci. 2021. PMID: 33526714 Free PMC article.
-
Kinetochores, cohesin, and DNA breaks: Controlling meiotic recombination within pericentromeres.Yeast. 2019 Mar;36(3):121-127. doi: 10.1002/yea.3366. Epub 2019 Feb 3. Yeast. 2019. PMID: 30625250 Free PMC article. Review.
-
The conserved histone variant H2A.Z illuminates meiotic recombination initiation.Curr Genet. 2018 Oct;64(5):1015-1019. doi: 10.1007/s00294-018-0825-9. Epub 2018 Mar 16. Curr Genet. 2018. PMID: 29549582 Review.
Cited by
-
Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation.PLoS Genet. 2015 May 20;11(5):e1005225. doi: 10.1371/journal.pgen.1005225. eCollection 2015 May. PLoS Genet. 2015. PMID: 25993311 Free PMC article.
-
Genomic Structure and Molecular Characterization of Toll-like Receptors in Black Scraper Thamnaconus Modestus and Their Expression Response to Two Types of Pathogens.Mar Biotechnol (NY). 2023 Oct;25(5):800-814. doi: 10.1007/s10126-023-10241-4. Epub 2023 Aug 11. Mar Biotechnol (NY). 2023. PMID: 37566262
-
Quantitative Genome-Wide Measurements of Meiotic DNA Double-Strand Breaks and Protein Binding in S. pombe.Methods Mol Biol. 2017;1471:25-49. doi: 10.1007/978-1-4939-6340-9_2. Methods Mol Biol. 2017. PMID: 28349389 Free PMC article.
-
Pfh1 Is an Accessory Replicative Helicase that Interacts with the Replisome to Facilitate Fork Progression and Preserve Genome Integrity.PLoS Genet. 2016 Sep 9;12(9):e1006238. doi: 10.1371/journal.pgen.1006238. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27611590 Free PMC article.
-
Differentiated function and localisation of SPO11-1 and PRD3 on the chromosome axis during meiotic DSB formation in Arabidopsis thaliana.PLoS Genet. 2022 Jul 20;18(7):e1010298. doi: 10.1371/journal.pgen.1010298. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35857772 Free PMC article.
References
-
- Bellani MA, Boateng KA, McLeod D, Camerini-Otero RD. The expression profile of the major mouse SPO11 isoforms indicates that SPO11β introduces double strand breaks and suggests that SPO11α has an additional role in prophase in both spermatocytes and oocytes. Mol Cell Biol. 2010;30:4391–4403. - PMC - PubMed
-
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

