Tcf15 primes pluripotent cells for differentiation
- PMID: 23395635
- PMCID: PMC3607254
- DOI: 10.1016/j.celrep.2013.01.017
Tcf15 primes pluripotent cells for differentiation
Abstract
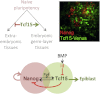
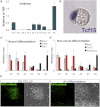
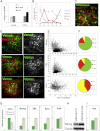
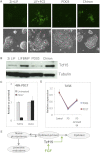
The events that prime pluripotent cells for differentiation are not well understood. Inhibitor of DNA binding/differentiation (Id) proteins, which are inhibitors of basic helix-loop-helix (bHLH) transcription factor activity, contribute to pluripotency by blocking sequential transitions toward differentiation. Using yeast-two-hybrid screens, we have identified Id-regulated transcription factors that are expressed in embryonic stem cells (ESCs). One of these, Tcf15, is also expressed in the embryonic day 4.5 embryo and is specifically associated with a novel subpopulation of primed ESCs. An Id-resistant form of Tcf15 rapidly downregulates Nanog and accelerates somatic lineage commitment. We propose that because Tcf15 can be held in an inactive state through Id activity, it may prime pluripotent cells for entry to somatic lineages upon downregulation of Id. We also find that Tcf15 expression is dependent on fibroblast growth factor (FGF) signaling, providing an explanation for how FGF can prime for differentiation without driving cells out of the pluripotent state.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Acampora D., Di Giovannantonio L.G., Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2012;140:43–55. - PubMed
-
- Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. - PubMed
-
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. - PubMed
-
- Brown D., Wagner D., Li X., Richardson J.A., Olson E.N. Dual role of the basic helix-loop-helix transcription factor scleraxis in mesoderm formation and chondrogenesis during mouse embryogenesis. Development. 1999;126:4317–4329. - PubMed
Supplemental References
-
- Dunning M.J., Smith M.L., Ritchie M.E., Tavaré S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. - PubMed
-
- Wettenhall J.M., Smyth G.K. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- G1100526/MRC_/Medical Research Council/United Kingdom
- G1001028/MRC_/Medical Research Council/United Kingdom
- G15381/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 079249/WT_/Wellcome Trust/United Kingdom
- BB/I006680/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

