Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection
- PMID: 23395983
- PMCID: PMC3622295
- DOI: 10.1093/gbe/evt021
Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection
Abstract
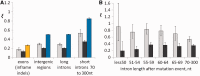
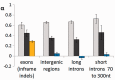
Insertions and deletions (collectively indels) obviously have a major impact on genome evolution. However, before large-scale data on indel polymorphism became available, it was difficult to estimate the strength of selection acting on indel mutations. Here, we analyze indel polymorphism and divergence in different compartments of the Drosophila melanogaster genome: exons, introns of different lengths, and intergenic regions. Data on low-frequency polymorphisms indicate that 0.036-0.039 short (1-30 nt) insertion mutations and 0.085-0.092 short deletion mutations, with mean lengths 3.23 and 4.78, respectively, occur per single-nucleotide substitution. The excess of short deletion over short insertion mutations implies that indel mutations of these lengths should lead to a loss of approximately 0.30 nt per single-nucleotide replacement. However, polymorphism and divergence data show that this deletion bias is almost completely compensated by selection: Negative selection is stronger against deletions, whereas insertions are more likely to be favored by positive selection. Among the inframe low-frequency polymorphic mutations in exons, long introns, and intergenic regions, selection prevents a larger fraction of deletions (80-87%, depending on the type of the compartment) than of insertions (70-82%) or single-nucleotide substitutions (49-73%), from reaching high frequencies. The corresponding fractions were the lowest in short introns: 66%, 47%, and 15%, respectively, consistent with the weakest selective constraint in them. The McDonald-Kreitman test shows that 32-46% of the deletions and 60-73% of the insertions that were fixed in the recent evolution of D. melanogaster are adaptive, whereas this fraction is only 0-29% for single-nucleotide substitutions.
Figures



Similar articles
-
The evolution of small insertions and deletions in the coding genes of Drosophila melanogaster.Mol Biol Evol. 2013 Dec;30(12):2699-708. doi: 10.1093/molbev/mst167. Epub 2013 Sep 26. Mol Biol Evol. 2013. PMID: 24077769
-
The correlation between intron length and recombination in drosophila. Dynamic equilibrium between mutational and selective forces.Genetics. 2000 Nov;156(3):1175-90. doi: 10.1093/genetics/156.3.1175. Genetics. 2000. PMID: 11063693 Free PMC article.
-
Short indels are subject to insertion-biased gene conversion.Evolution. 2013 Sep;67(9):2604-13. doi: 10.1111/evo.12129. Epub 2013 May 11. Evolution. 2013. PMID: 24033170
-
Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review.
-
Insertions and deletions in protein evolution and engineering.Biotechnol Adv. 2022 Nov;60:108010. doi: 10.1016/j.biotechadv.2022.108010. Epub 2022 Jun 20. Biotechnol Adv. 2022. PMID: 35738511 Review.
Cited by
-
Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster.Genetics. 2013 Aug;194(4):937-54. doi: 10.1534/genetics.113.151670. Epub 2013 Jun 3. Genetics. 2013. PMID: 23733788 Free PMC article.
-
In silico evolution of globular protein folds from random sequences.Proc Natl Acad Sci U S A. 2025 Jul 8;122(27):e2509015122. doi: 10.1073/pnas.2509015122. Epub 2025 Jun 30. Proc Natl Acad Sci U S A. 2025. PMID: 40587803 Free PMC article.
-
Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines.Genome Res. 2014 Jul;24(7):1193-208. doi: 10.1101/gr.171546.113. Epub 2014 Apr 8. Genome Res. 2014. PMID: 24714809 Free PMC article.
-
Molecular Population Genetics.Genetics. 2017 Mar;205(3):1003-1035. doi: 10.1534/genetics.116.196493. Genetics. 2017. PMID: 28270526 Free PMC article. Review.
-
The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences.BMC Genomics. 2013 Jul 15;14:476. doi: 10.1186/1471-2164-14-476. BMC Genomics. 2013. PMID: 23855885 Free PMC article.
References
-
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. - PubMed
-
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. - PubMed
-
- Blumenstiel JP, Hartl DL, Lozovsky ER. Patterns of insertion and deletion in contrasting chromatin domains. Mol Biol Evol. 2002;19:2211–2225. - PubMed
-
- Casillas S, Barbadilla A, Bergman CM. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol. 2007;24:2222–2234. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

