Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples
- PMID: 23396013
- PMCID: PMC3833702
- DOI: 10.1038/nbt.2514
Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples
Abstract
Detection of somatic point substitutions is a key step in characterizing the cancer genome. However, existing methods typically miss low-allelic-fraction mutations that occur in only a subset of the sequenced cells owing to either tumor heterogeneity or contamination by normal cells. Here we present MuTect, a method that applies a Bayesian classifier to detect somatic mutations with very low allele fractions, requiring only a few supporting reads, followed by carefully tuned filters that ensure high specificity. We also describe benchmarking approaches that use real, rather than simulated, sequencing data to evaluate the sensitivity and specificity as a function of sequencing depth, base quality and allelic fraction. Compared with other methods, MuTect has higher sensitivity with similar specificity, especially for mutations with allelic fractions as low as 0.1 and below, making MuTect particularly useful for studying cancer subclones and their evolution in standard exome and genome sequencing data.
Figures

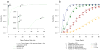

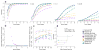
Comment in
-
Bioinformatics: Tumors have their differences.Nat Methods. 2013 Apr;10(4):282-3. doi: 10.1038/nmeth.2426. Nat Methods. 2013. PMID: 23653923 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

