Long-term dynamics of CA1 hippocampal place codes
- PMID: 23396101
- PMCID: PMC3784308
- DOI: 10.1038/nn.3329
Long-term dynamics of CA1 hippocampal place codes
Abstract
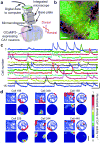
Using Ca(2+) imaging in freely behaving mice that repeatedly explored a familiar environment, we tracked thousands of CA1 pyramidal cells' place fields over weeks. Place coding was dynamic, as each day the ensemble representation of this environment involved a unique subset of cells. However, cells in the ∼15-25% overlap between any two of these subsets retained the same place fields, which sufficed to preserve an accurate spatial representation across weeks.
Figures


References
-
- O'Keefe JNL. The Hippocampus as a Cognitive Map. Clarendon Oxford; 1978.
-
- Leutgeb S, et al. Independent codes for spatial and episodic memory in hippocampal neuronal ensembles. Science. 2005;309:619–623. 309/5734/619 [pii] 10.1126/science.1114037. - PubMed
-
- Buzsaki G. Large-scale recording of neuronal ensembles. Nat Neurosci. 2004;7:446–451. 10.1038/nn1233 nn1233 [pii] - PubMed
-
- Thompson LT, Best PJ. Long-term stability of the place-field activity of single units recorded from the dorsal hippocampus of freely behaving rats. Brain Res. 1990;509:299–308. 0006-8993(90)90555-P [pii] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

