Effects of the peptide pheromone plantaricin A and cocultivation with Lactobacillus sanfranciscensis DPPMA174 on the exoproteome and the adhesion capacity of Lactobacillus plantarum DC400
- PMID: 23396346
- PMCID: PMC3623163
- DOI: 10.1128/AEM.03625-12
Effects of the peptide pheromone plantaricin A and cocultivation with Lactobacillus sanfranciscensis DPPMA174 on the exoproteome and the adhesion capacity of Lactobacillus plantarum DC400
Abstract
This study aimed at investigating the extracellular and cell wall-associated proteins (exoproteome) of Lactobacillus plantarum DC400 when cultivated on modified chemically defined medium (CDM) supplemented with the chemically synthesized pheromone plantaricin A (PlnA) or cocultured with L. plantarum DPPMA20 or Lactobacillus sanfranciscensis DPPMA174. Compared to monoculture, two-dimensional gel electrophoresis (2-DE) analysis showed that the exoproteome of L. plantarum DC400 was affected by PlnA and cocultivation with strains DPPMA20 and, especially, DPPMA174. The highest similarity of the 2-DE maps was found between DC400 cells cultivated in monoculture and in coculture with strain DPPMA20. Almost all extracellular proteins (22 spots) and cell wall-associated proteins (40 spots) which showed decreased or increased levels of synthesis during growth in CDM supplemented with PlnA and/or in coculture with strain DPPMA20 or DPPMA174 were identified. On the basis of the sequences in the Kyoto Encyclopedia of Genes and Genomes database, changes to the exoproteome concerned proteins involved in quorum sensing (QS), the transport system, stress response, carbohydrate metabolism and glycolysis, oxidation/reduction processes, the proteolytic system, amino acid metabolism, cell wall and catabolic processes, and cell shape, growth, and division. Cultivation with PlnA and cocultivation with strains DPPMA20 and, especially, DPMMA174 markedly increased the capacity of L. plantarum DC400 to form biofilms, to adhere to human Caco-2 cells, and to prevent the adhesion of potential intestinal pathogens. These phenotypic traits were in part related to oversynthesized moonlighting proteins (e.g., DnaK and GroEL, pyruvate kinase, enolase, and glyceraldehyde-3-phosphate dehydrogenase) in response to QS mechanisms and interaction with L. plantarum DPPMA20 and, especially, L. sanfranciscensis DPPMA174.
Figures

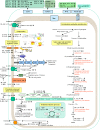
 or
or  ) or in coculture with L. plantarum DPPMA20 (
) or in coculture with L. plantarum DPPMA20 ( or
or  ) or Lactobacillus sanfranciscensis DPPMA174 (
) or Lactobacillus sanfranciscensis DPPMA174 ( or
or  ). Compared to the amounts of proteins in monoculture, the absence of changes in the amounts of proteins (average of three replicates) is represented with a minus sign. Protein names and spot numbers correspond to those in Tables 1 and 2. -E, extracellular proteins.
). Compared to the amounts of proteins in monoculture, the absence of changes in the amounts of proteins (average of three replicates) is represented with a minus sign. Protein names and spot numbers correspond to those in Tables 1 and 2. -E, extracellular proteins.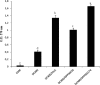
Similar articles
-
Quorum sensing in sourdough Lactobacillus plantarum DC400: induction of plantaricin A (PlnA) under co-cultivation with other lactic acid bacteria and effect of PlnA on bacterial and Caco-2 cells.Proteomics. 2010 Jun;10(11):2175-90. doi: 10.1002/pmic.200900565. Proteomics. 2010. PMID: 20354993
-
Molecular adaptation of sourdough Lactobacillus plantarum DC400 under co-cultivation with other lactobacilli.Res Microbiol. 2009 Jun;160(5):358-66. doi: 10.1016/j.resmic.2009.04.006. Epub 2009 May 13. Res Microbiol. 2009. PMID: 19446023
-
Plantaricin A synthesized by Lactobacillus plantarum induces in vitro proliferation and migration of human keratinocytes and increases the expression of TGF-β1, FGF7, VEGF-A and IL-8 genes.Peptides. 2011 Sep;32(9):1815-24. doi: 10.1016/j.peptides.2011.07.004. Epub 2011 Jul 14. Peptides. 2011. PMID: 21782870
-
Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1.Microbiology (Reading). 2007 Dec;153(Pt 12):3939-3947. doi: 10.1099/mic.0.2007/012831-0. Microbiology (Reading). 2007. PMID: 18048908 Review.
-
An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum.Peptides. 2009 Aug;30(8):1562-74. doi: 10.1016/j.peptides.2009.05.014. Epub 2009 May 22. Peptides. 2009. PMID: 19465075 Review.
Cited by
-
Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections.Sensors (Basel). 2013 Apr 18;13(4):5130-66. doi: 10.3390/s130405130. Sensors (Basel). 2013. PMID: 23598501 Free PMC article. Review.
-
Identification and Characterization of Probiotic Lactiplantibacillus plantarum BI-59.1 Isolated from tejuino and Its Capacity to Produce Biofilms.Curr Microbiol. 2023 May 19;80(7):220. doi: 10.1007/s00284-023-03319-8. Curr Microbiol. 2023. PMID: 37204589
-
Fighting biofilms with lantibiotics and other groups of bacteriocins.NPJ Biofilms Microbiomes. 2018 Apr 19;4:9. doi: 10.1038/s41522-018-0053-6. eCollection 2018. NPJ Biofilms Microbiomes. 2018. PMID: 29707229 Free PMC article. Review.
-
Whole-Genome Sequencing, Phylogenetic and Genomic Analysis of Lactiplantibacillus pentosus L33, a Potential Probiotic Strain Isolated From Fermented Sausages.Front Microbiol. 2021 Oct 26;12:746659. doi: 10.3389/fmicb.2021.746659. eCollection 2021. Front Microbiol. 2021. PMID: 34764945 Free PMC article.
-
Bacteriocin production enhancing mechanism of Lactiplantibacillus paraplantarum RX-8 response to Wickerhamomyces anomalus Y-5 by transcriptomic and proteomic analyses.Front Microbiol. 2023 Feb 24;14:1111516. doi: 10.3389/fmicb.2023.1111516. eCollection 2023. Front Microbiol. 2023. PMID: 36910197 Free PMC article.
References
-
- Zhou M, Theunissen D, Wels M, Siezen RJ. 2010. LAB-secretome: a genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of lactic acid bacteria. BMC Genomics 11:651 doi:10.1186/1471-2164-11-651 - DOI - PMC - PubMed
-
- Roos S, Jonsson H. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433–442 - PubMed
-
- Quadri LE. 2002. Regulation of antimicrobial peptide production by autoinducer-mediated quorum sensing in lactic acid bacteria. Antonie Van Leeuwenhoek 82:133–145 - PubMed
-
- de Vos WM. 2011. Systems solutions by lactic acid bacteria: from paradigms to practice. Microb. Cell Fact. 10(Suppl 1):S2 doi:10.1186/1475-2859-10-S1-S2 - DOI - PMC - PubMed
-
- De Angelis M, Gobbetti M. 2011. Lactic acid bacteria. Lactobacillus spp.: general characteristics, p 78–90 In Fuquay JW, Fox PF, McSweeney PLH. (ed), Encyclopedia of dairy sciences, 2nd ed, vol 3 Academic Press, San Diego, CA
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

