Proteomics of genetically engineered mouse mammary tumors identifies fatty acid metabolism members as potential predictive markers for cisplatin resistance
- PMID: 23397111
- PMCID: PMC3650342
- DOI: 10.1074/mcp.M112.024182
Proteomics of genetically engineered mouse mammary tumors identifies fatty acid metabolism members as potential predictive markers for cisplatin resistance
Abstract
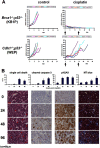
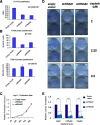
In contrast to various signatures that predict the prognosis of breast cancer patients, markers that predict chemotherapy response are still elusive. To detect such predictive biomarkers, we investigated early changes in protein expression using two mouse models for distinct breast cancer subtypes who have a differential knock-out status for the breast cancer 1, early onset (Brca1) gene. The proteome of cisplatin-sensitive BRCA1-deficient mammary tumors was compared with that of cisplatin-resistant mammary tumors resembling pleomorphic invasive lobular carcinoma. The analyses were performed 24 h after administration of the maximum tolerable dose of cisplatin. At this time point, drug-sensitive BRCA1-deficient tumors showed DNA damage, but cells were largely viable. By applying paired statistics and quantitative filtering, we identified highly discriminatory markers for the sensitive and resistant model. Proteins up-regulated in the sensitive model are involved in centrosome organization, chromosome condensation, homology-directed DNA repair, and nucleotide metabolism. Major discriminatory markers that were up-regulated in the resistant model were predominantly involved in fatty acid metabolism, such as fatty-acid synthase. Specific inhibition of fatty-acid synthase sensitized resistant cells to cisplatin. Our data suggest that exploring the functional link between the DNA damage response and cancer metabolism shortly after the initial treatment may be a useful strategy to predict the efficacy of cisplatin.
Figures



References
-
- Borst P., Wessels L. (2010) Do predictive signatures really predict response to cancer chemotherapy? Cell Cycle 9, 4836–4840 - PubMed
-
- Liu X., Holstege H., van der Gulden H., Treur-Mulder M., Zevenhoven J., Velds A., Kerkhoven R. M., van Vliet M. H., Wessels L. F., Peterse J. L., Berns A., Jonkers J. (2007) Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12111–12116 - PMC - PubMed
-
- Rottenberg S., Nygren A. O., Pajic M., van Leeuwen F. W., van der Heijden I., van de Wetering K., Liu X., de Visser K. E., Gilhuijs K. G., van Tellingen O., Schouten J. P., Jonkers J., Borst P. (2007) Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12117–12122 - PMC - PubMed
-
- Vollebergh M. A., Lips E. H., Nederlof P. M., Wessels L. F., Schmidt M. K., van Beers E. H., Cornelissen S., Holtkamp M., Froklage F. E., de Vries E. G., Schrama J. G., Wesseling J., van, d. V., van T. H., de B. M., Hauptmann M., Rodenhuis S., Linn S. C. (2010) An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann. Oncol., 22, 1561–1570 - PMC - PubMed
-
- Silver D. P., Richardson A. L., Eklund A. C., Wang Z. C., Szallasi Z., Li Q., Juul N., Leong C. O., Calogrias D., Buraimoh A., Fatima A., Gelman R. S., Ryan P. D., Tung N. M., De Nicolo A., Ganesan S., Miron A., Colin C., Sgroi D. C., Ellisen L. W., Winer E. P., Garber J. E. (2010) Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. 28, 1145–1153 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

