The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability
- PMID: 23398456
- PMCID: PMC3632086
- DOI: 10.1042/BJ20130026
The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability
Abstract
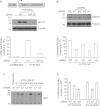
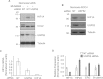
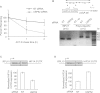
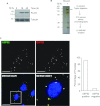
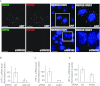
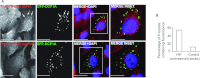
HIF1A (hypoxia-inducible factor 1α) is the master regulator of the cellular response to hypoxia and is implicated in cancer progression. Whereas the regulation of HIF1A protein in response to oxygen is well characterized, less is known about the fate of HIF1A mRNA. In the present study, we have identified the pseudo-DUB (deubiquitinating enzyme)/deadenylase USP52 (ubiquitin-specific protease 52)/PAN2 [poly(A) nuclease 2] as an important regulator of the HIF1A-mediated hypoxic response. Depletion of USP52 reduced HIF1A mRNA and protein levels and resulted in reduced expression of HIF1A-regulated hypoxic targets due to a 3'-UTR (untranslated region)-dependent poly(A)-tail-length-independent destabilization in HIF1A mRNA. MS analysis revealed an association of USP52 with several P-body (processing body) components and we confirmed further that USP52 protein and HIF1A mRNA co-localized with cytoplasmic P-bodies. Importantly, P-body dispersal by knockdown of GW182 or LSM1 resulted in a reduction of HIF1A mRNA levels. These data uncover a novel role for P-bodies in regulating HIF1A mRNA stability, and demonstrate that USP52 is a key component of P-bodies required to prevent HIF1A mRNA degradation.
Figures

References
-
- Kenneth N. S., Rocha S. Regulation of gene expression by hypoxia. Biochem. J. 2008;414:19–29. - PubMed
-
- Salceda S., Caro J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin–proteasome system under normoxic conditions: its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 1997;272:22642–22647. - PubMed
-
- Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. - PubMed
-
- Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O’Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

