Combined ChIP-Seq and transcriptome analysis identifies AP-1/JunD as a primary regulator of oxidative stress and IL-1β synthesis in macrophages
- PMID: 23398888
- PMCID: PMC3608227
- DOI: 10.1186/1471-2164-14-92
Combined ChIP-Seq and transcriptome analysis identifies AP-1/JunD as a primary regulator of oxidative stress and IL-1β synthesis in macrophages
Abstract
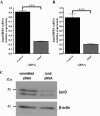
Background: The oxidative burst is one of the major antimicrobial mechanisms adopted by macrophages. The WKY rat strain is uniquely susceptible to experimentally induced macrophage-dependent crescentic glomerulonephritis (Crgn). We previously identified the AP-1 transcription factor JunD as a determinant of macrophage activation in WKY bone marrow-derived macrophages (BMDMs). JunD is over-expressed in WKY BMDMs and its silencing reduces Fc receptor-mediated oxidative burst in these cells.
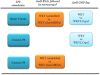
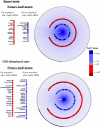
Results: Here we combined Jund RNA interference with microarray analyses alongside ChIP-sequencing (ChIP-Seq) analyses in WKY BMDMs to investigate JunD-mediated control of macrophage activation in basal and lipopolysaccharide (LPS) stimulated cells. Microarray analysis following Jund silencing showed that Jund activates and represses gene expression with marked differential expression (>3 fold) for genes linked with oxidative stress and IL-1β expression. These results were complemented by comparing whole genome expression in WKY BMDMs with Jund congenic strain (WKY.LCrgn2) BMDMs which express lower levels of JunD. ChIP-Seq analyses demonstrated that the increased expression of JunD resulted in an increased number of binding events in WKY BMDMs compared to WKY.LCrgn2 BMDMs. Combined ChIP-Seq and microarray analysis revealed a set of primary JunD-targets through which JunD exerts its effect on oxidative stress and IL-1β synthesis in basal and LPS-stimulated macrophages.
Conclusions: These findings demonstrate how genetically determined levels of a transcription factor affect its binding sites in primary cells and identify JunD as a key regulator of oxidative stress and IL-1β synthesis in primary macrophages, which may play a role in susceptibility to Crgn.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

