Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells
- PMID: 23399548
- PMCID: PMC3617340
- DOI: 10.1074/mcp.O112.021972
Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells
Abstract
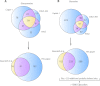
Characterizing protein GalNAc-type O-glycosylation has long been a major challenge, and as a result, our understanding of this glycoproteome is particularly poor. Recently, we presented a novel strategy for high throughput identification of O-GalNAc glycosites using zinc finger nuclease gene-engineered "SimpleCell" lines producing homogeneous truncated O-glycosylation. Total lysates of cells were trypsinized and subjected to lectin affinity chromatography enrichment, followed by identification of GalNAc O-glycopeptides by nLC-MS/MS, with electron transfer dissociation employed to specify sites of O-glycosylation. Here, we demonstrate a substantial improvement in the SimpleCell strategy by including an additional stage of lectin affinity chromatography on secreted glycoproteins from culture media (secretome) and by incorporating pre-fractionation of affinity-enriched glycopeptides via IEF before nLC-MS/MS. We applied these improvements to three human SimpleCells studied previously, and each yielded a substantial increase in the number of O-glycoproteins and O-glycosites identified. We found that analysis of the secretome was an important independent factor for increasing identifications, suggesting that further substantial improvements can also be sought through analysis of subcellular organelle fractions. In addition, we uncovered a substantial nonoverlapping set of O-glycoproteins and O-glycosites using an alternative protease digestion (chymotrypsin). In total, the improvements led to identification of 259 glycoproteins, of which 152 (59%) were novel compared with our previous strategy using the same three cell lines. With respect to individual glycosites, we identified a total of 856 sites, of which 508 (59%) were novel compared with our previous strategy; this includes four new identifications of O-GalNAc attached to tyrosine. Furthermore, we uncovered ≈ 220 O-glycosites wherein the peptides were clearly identified, but the glycosites could not be unambiguously assigned to specific positions. The improved strategy should greatly facilitate high throughput characterization of the human GalNAc-type O-glycoproteome as well as be applicable to analysis of other O-glycoproteomes.
Figures





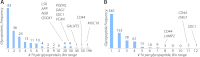
References
-
- Lowe J. B., Marth J. D. (2003) A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72, 643–691 - PubMed
-
- Gill D. J., Clausen H., Bard F. (2011) Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21, 149–158 - PubMed
-
- Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., Nilsson J. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid β-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 108, 11848–11853 - PMC - PubMed
-
- Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

