Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology
- PMID: 23399914
- PMCID: PMC3807661
- DOI: 10.1038/mp.2013.1
Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology
Abstract
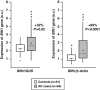
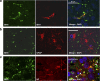
Genome-wide association studies (GWAS) have identified a region upstream the BIN1 gene as the most important genetic susceptibility locus in Alzheimer's disease (AD) after APOE. We report that BIN1 transcript levels were increased in AD brains and identified a novel 3 bp insertion allele ∼28 kb upstream of BIN1, which increased (i) transcriptional activity in vitro, (ii) BIN1 expression levels in human brain and (iii) AD risk in three independent case-control cohorts (Meta-analysed Odds ratio of 1.20 (1.14-1.26) (P=3.8 × 10(-11))). Interestingly, decreased expression of the Drosophila BIN1 ortholog Amph suppressed Tau-mediated neurotoxicity in three different assays. Accordingly, Tau and BIN1 colocalized and interacted in human neuroblastoma cells and in mouse brain. Finally, the 3 bp insertion was associated with Tau but not Amyloid loads in AD brains. We propose that BIN1 mediates AD risk by modulating Tau pathology.
Figures



References
-
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. - PubMed
-
- Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

