Circulating microRNA expression profiles associated with systemic lupus erythematosus
- PMID: 23401079
- PMCID: PMC6662589
- DOI: 10.1002/art.37890
Circulating microRNA expression profiles associated with systemic lupus erythematosus
Abstract
Objective: To evaluate the specificity of expression patterns of cell-free circulating microRNAs (miRNAs) in systemic lupus erythematosus (SLE).
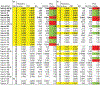
Methods: Total RNA was purified from plasma, and 45 different specific, mature miRNAs were determined using quantitative reverse transcription-polymerase chain reaction assays. A total of 409 plasma samples were obtained from 364 different patients with SLE, healthy control subjects, and control subjects with other autoimmune diseases. The results in the primary cohort of 62 patients with SLE and 29 healthy control subjects were validated in 2 independent cohorts: a validation cohort comprising 68 patients with SLE and 68 healthy control subjects, and a disease control cohort comprising 20 patients with SLE (19 of whom were from the other validation cohort), 46 healthy control subjects, 38 patients with vasculitis, 18 patients with rheumatoid arthritis, and 20 immunosuppressed patients.
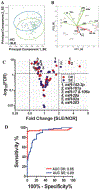
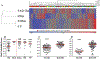
Results: Seven miRNAs were statistically significantly differentially expressed in plasma from patients with SLE. The expression of miRNA-142-3p (miR-142-3p) and miR-181a was increased, and the expression of miR-106a, miR-17, miR-20a, miR-203, and miR-92a was decreased. In addition, the expression of miR-342-3p, miR-223, and miR-20a was significantly decreased in SLE patients with active nephritis. A predictive model for SLE based on 2 or 4 miRNAs differentiated patients with SLE from control subjects (76% accuracy) when validated independently (P < 2 × 10(-9) ). Use of the 4-miRNA model provided highly significant differentiation between the SLE group and disease controls, except for those with vasculitis.
Conclusion: Circulating miRNAs are systematically altered in SLE. A 4-miRNA signature was diagnostic of SLE, and a specific subset of miRNA profiles was associated with nephritis. All of the signature miRNAs target genes in the transforming growth factor β signaling pathways. Other targets include regulation of apoptosis, cytokine-cytokine receptors, T cell development, and cytoskeletal organization. These findings highlight possible dysregulated pathways in SLE and suggest that circulating miRNA patterns distinguish SLE from other immunoinflammatory phenotypes.
Copyright © 2013 by the American College of Rheumatology.
Figures

References
-
- Ronnblom L, Alm GV, Eloranta ML. Type I interferon and lupus. Curr Opin Rheumatol 2009;21:471–7. - PubMed
-
- Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol 2004;16:801–7. - PubMed
-
- Eloranta ML, Lovgren T, Finke D, Mathsson L, Ronnelid J, Kastner B, et al. Regulation of the interferon-α production induced by RNA-containing immune complexes in plasmacytoid dendritic cells. Arthritis Rheum 2009;60:2418–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

