Bridging the genotype-phenotype gap: what does it take?
- PMID: 23401613
- PMCID: PMC3634519
- DOI: 10.1113/jphysiol.2012.248864
Bridging the genotype-phenotype gap: what does it take?
Abstract
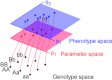
The genotype-phenotype map (GP map) concept applies to any time point in the ontogeny of a living system. It is the outcome of very complex dynamics that include environmental effects, and bridging the genotype-phenotype gap is synonymous with understanding these dynamics. The context for this understanding is physiology, and the disciplinary goals of physiology do indeed demand the physiological community to seek this understanding. We claim that this task is beyond reach without use of mathematical models that bind together genetic and phenotypic data in a causally cohesive way. We provide illustrations of such causally cohesive genotype-phenotype models where the phenotypes span from gene expression profiles to development of whole organs. Bridging the genotype-phenotype gap also demands that large-scale biological ('omics') data and associated bioinformatics resources be more effectively integrated with computational physiology than is currently the case. A third major element is the need for developing a phenomics technology way beyond current state of the art, and we advocate the establishment of a Human Phenome Programme solidly grounded on biophysically based mathematical descriptions of human physiology.
Figures


References
-
- Alberch P. From genes to phenotype: dynamical systems and evolvability. Genetica. 1991;84:5–11. - PubMed
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL, USA: Chapman and Hall/CRC; 2006.
-
- Altinok A, Lévi F, Goldbeter A. A cell cycle automaton model for probing circadian patterns of anticancer drug delivery. Adv Drug Deliv Rev. 2007;59:1036–1053. - PubMed
-
- Aster RC, Borchers B, Thurber CH. Parameter Estimation and Inverse Problems, Second Edition. 2nd edn. New York: Academic Press; 2012.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

