Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III
- PMID: 23401852
- PMCID: PMC3624256
- DOI: 10.1128/MCB.01733-12
Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III
Abstract
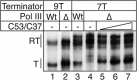
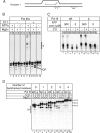
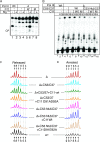
Transcription termination by RNA polymerase (Pol) III serves multiple purposes; it delimits interference with downstream genes, forms 3' oligo(U) binding sites for the posttranscriptional processing factor, La protein, and resets the polymerase complex for reinitiation. Although an interplay of several Pol III subunits is known to collectively control these activities, how they affect molecular function of the active center during termination is incompletely understood. We have approached this using immobilized Pol III-nucleic acid scaffolds to examine the two major components of termination, transcription pausing and RNA release. This allowed us to distinguish two mechanisms of termination by isolated Saccharomyces cerevisiae Pol III. A core mechanism can operate in the absence of C53/37 and C11 subunits but requires synthesis of 8 or more 3' U nucleotides, apparently reflecting inherent sensitivity to an oligo(rU·dA) hybrid that is the termination signal proper. The holoenzyme mechanism requires fewer U nucleotides but uses C53/37 and C11 to slow elongation and prevent terminator arrest. N-terminal truncation of C53 or point mutations that disable the cleavage activity of C11 impair their antiarrest activities. The data are consistent with a model in which C53, C37, and C11 activities are functionally integrated with the active center of Pol III during termination.
Figures



References
-
- Roeder RG, Rutter WJ. 1969. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 224:234–237 - PubMed
-
- Carter R, Drouin G. 2010. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 27:1035–1043 - PubMed
-
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ. 2012. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337:599–602 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
