P19ARF and RasV¹² offer opposing regulation of DHX33 translation to dictate tumor cell fate
- PMID: 23401854
- PMCID: PMC3624250
- DOI: 10.1128/MCB.01220-12
P19ARF and RasV¹² offer opposing regulation of DHX33 translation to dictate tumor cell fate
Abstract
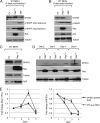
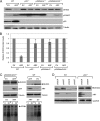
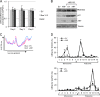
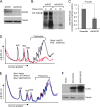
DHX33 is a pivotal DEAH-box RNA helicase in the multistep process of RNA polymerase I-directed transcription of the ribosomal DNA locus. We explored the regulation of DHX33 expression by Ras(V12) and ARF to determine DHX33's role in sensing these opposing signals to regulate ribosome biogenesis. In wild-type primary fibroblasts, Ras(V12) infection induced a transient increase in DHX33 protein level, as well as an rRNA transcriptional rate that was eventually suppressed by a delayed activation of the ARF/p53 pathway. DHX33 expression was exclusively controlled at the level of translation. ARF caused a dramatic reduction in polysome-associated DHX33 mRNAs, while Ras(V12) led to a complete shift of existing DHX33 mRNAs to actively translating polysomes. The translation of DHX33 by Ras(V12) was sensitive to inhibitors of phosphatidylinositol 3-kinase, mTOR, and mitogen-activated protein and was pivotal for enhanced rRNA transcription and enhanced overall cellular protein translation. In addition, DHX33 knockdown abolished Ras(V12)-induced rRNA transcription and protein translation and prevented both the in vitro and in vivo transforming properties of oncogenic Ras(V12). Our results directly implicate DHX33 as a crucial player in establishing rRNA synthesis rates in the face of Ras(V12) or ARF signals, adjusting ribosome biogenesis to match the appropriate growth or antigrowth signals.
Figures

Similar articles
-
Identification of DHX33 as a mediator of rRNA synthesis and cell growth.Mol Cell Biol. 2011 Dec;31(23):4676-91. doi: 10.1128/MCB.05832-11. Epub 2011 Sep 19. Mol Cell Biol. 2011. PMID: 21930779 Free PMC article.
-
The ARF tumor-suppressor controls Drosha translation to prevent Ras-driven transformation.Oncogene. 2014 Jan 16;33(3):300-7. doi: 10.1038/onc.2012.601. Epub 2013 Jan 14. Oncogene. 2014. PMID: 23318441 Free PMC article.
-
Depletion of ERK2 but not ERK1 abrogates oncogenic Ras-induced senescence.Cell Signal. 2013 Dec;25(12):2540-7. doi: 10.1016/j.cellsig.2013.08.014. Epub 2013 Aug 30. Cell Signal. 2013. PMID: 23993963
-
ARF tumor suppression in the nucleolus.Biochim Biophys Acta. 2014 Jun;1842(6):831-9. doi: 10.1016/j.bbadis.2014.01.016. Epub 2014 Feb 10. Biochim Biophys Acta. 2014. PMID: 24525025 Review.
-
Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis.Biochim Biophys Acta. 2014 Jun;1842(6):817-30. doi: 10.1016/j.bbadis.2013.08.014. Epub 2013 Oct 26. Biochim Biophys Acta. 2014. PMID: 24514102 Review.
Cited by
-
The DHX33 RNA Helicase Promotes mRNA Translation Initiation.Mol Cell Biol. 2015 Sep 1;35(17):2918-31. doi: 10.1128/MCB.00315-15. Epub 2015 Jun 22. Mol Cell Biol. 2015. PMID: 26100019 Free PMC article.
-
Targeting RNA helicase DHX33 blocks Ras-driven lung tumorigenesis in vivo.Cancer Sci. 2020 Oct;111(10):3564-3575. doi: 10.1111/cas.14601. Epub 2020 Aug 24. Cancer Sci. 2020. PMID: 32767810 Free PMC article.
-
miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33.Mol Oncol. 2016 Dec;10(10):1532-1541. doi: 10.1016/j.molonc.2016.09.001. Epub 2016 Sep 20. Mol Oncol. 2016. PMID: 27693040 Free PMC article.
-
Reconstruction of gene regulatory networks reveals chromatin remodelers and key transcription factors in tumorigenesis.Genome Med. 2016 May 19;8(1):57. doi: 10.1186/s13073-016-0310-3. Genome Med. 2016. PMID: 27198694 Free PMC article.
-
Depletion of the cisplatin targeted HMGB-box factor UBF selectively induces p53-independent apoptotic death in transformed cells.Oncotarget. 2015 Sep 29;6(29):27519-36. doi: 10.18632/oncotarget.4823. Oncotarget. 2015. PMID: 26317157 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
