The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination
- PMID: 23401855
- PMCID: PMC3624244
- DOI: 10.1128/MCB.01503-12
The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination
Abstract
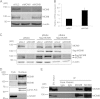
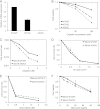
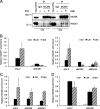
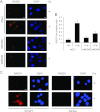
The minichromosome maintenance protein homologs MCM8 and MCM9 have previously been implicated in DNA replication elongation and prereplication complex (pre-RC) formation, respectively. We found that MCM8 and MCM9 physically associate with each other and that MCM8 is required for the stability of MCM9 protein in mammalian cells. Depletion of MCM8 or MCM9 in human cancer cells or the loss of function MCM9 mutation in mouse embryo fibroblasts sensitizes cells to the DNA interstrand cross-linking (ICL) agent cisplatin. Consistent with a role in the repair of ICLs by homologous recombination (HR), knockdown of MCM8 or MCM9 significantly reduces HR repair efficiency. Chromatin immunoprecipitation analysis using human DR-GFP cells or Xenopus egg extract demonstrated that MCM8 and MCM9 proteins are rapidly recruited to DNA damage sites and promote RAD51 recruitment. Thus, these two metazoan-specific MCM homologs are new components of HR and may represent novel targets for treating cancer in combination with DNA cross-linking agents.
Figures



References
-
- Scharer OD. 2005. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem 6:27–32 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
