Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency
- PMID: 23402340
- PMCID: PMC3621168
- DOI: 10.1186/1471-2229-13-26
Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency
Abstract
Background: TIFY is a large plant-specific transcription factor gene family. A subgroup of TIFY genes named JAZ (Jasmonate-ZIM domain) has been identified as repressors of jasmonate (JA)-regulated transcription in Arabidopsis and other plants. JA signaling is involved in many aspects of plant growth/development and in defense responses to biotic and abiotic stresses. Here, we identified the TIFY genes (designated PvTIFY) from the legume common bean (Phaseolus vulgaris) and functionally characterized PvTIFY10C as a transcriptional regulator.
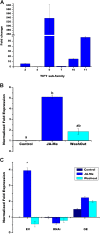
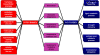
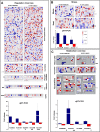
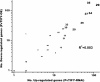
Results: Nineteen genes from the PvTIFY gene family were identified through whole-genome sequence analysis. Most of these were induced upon methyl-JA elicitation. We selected PvTIFY10C as a representative JA-responsive PvTIFY gene for further functional analysis. Transcriptome analysis via microarray hybridization using the newly designed Bean Custom Array 90 K was performed on transgenic roots of composite plants with modulated (RNAi-silencing or over-expression) PvTIFY10C gene expression. Data were interpreted using Gene Ontology and MapMan adapted to common bean. Microarray differential gene expression data were validated by real-time qRT-PCR expression analysis. Comparative global gene expression analysis revealed opposite regulatory changes in processes such as RNA and protein regulation, stress responses and metabolism in PvTIFY10C silenced vs. over-expressing roots. These data point to transcript reprogramming (mainly repression) orchestrated by PvTIFY10C. In addition, we found that several PvTIFY genes, as well as genes from the JA biosynthetic pathway, responded to P-deficiency. Relevant P-responsive genes that participate in carbon metabolic pathways, cell wall synthesis, lipid metabolism, transport, DNA, RNA and protein regulation, and signaling were oppositely-regulated in control vs. PvTIFY10C-silenced roots of composite plants under P-stress. These data indicate that PvTIFY10C regulates, directly or indirectly, the expression of some P-responsive genes; this process could be mediated by JA-signaling.
Conclusion: Our work contributes to the functional characterization of PvTIFY transcriptional regulators in common bean, an agronomically important legume. Members from the large PvTIFY gene family are important global transcriptional regulators that could participate as repressors in the JA signaling pathway. In addition, we propose that the JA-signaling pathway involving PvTIFY genes might play a role in regulating the plant response/adaptation to P-starvation.
Figures



References
-
- Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus spp.) – model food legumes. Plant Soil. 2003;252:55–128.
-
- Phaseolus vulgaris v1.0. DOE-JGI and USDA-NIFA; http://www.phytozome.net/commonbean.
-
- Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, Wu HC, Lara M, Town CD, Kopka J, Udvardi MK, Vance CP. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 2007;144:752–767. doi: 10.1104/pp.107.096958. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

