Open source tools for management and archiving of digital microscopy data to allow integration with patient pathology and treatment information
- PMID: 23402499
- PMCID: PMC3575263
- DOI: 10.1186/1746-1596-8-22
Open source tools for management and archiving of digital microscopy data to allow integration with patient pathology and treatment information
Abstract
Background: Virtual microscopy includes digitisation of histology slides and the use of computer technologies for complex investigation of diseases such as cancer. However, automated image analysis, or website publishing of such digital images, is hampered by their large file sizes.
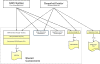
Results: We have developed two Java based open source tools: Snapshot Creator and NDPI-Splitter. Snapshot Creator converts a portion of a large digital slide into a desired quality JPEG image. The image is linked to the patient's clinical and treatment information in a customised open source cancer data management software (Caisis) in use at the Australian Breast Cancer Tissue Bank (ABCTB) and then published on the ABCTB website (http://www.abctb.org.au) using Deep Zoom open source technology. Using the ABCTB online search engine, digital images can be searched by defining various criteria such as cancer type, or biomarkers expressed. NDPI-Splitter splits a large image file into smaller sections of TIFF images so that they can be easily analysed by image analysis software such as Metamorph or Matlab. NDPI-Splitter also has the capacity to filter out empty images.
Conclusions: Snapshot Creator and NDPI-Splitter are novel open source Java tools. They convert digital slides into files of smaller size for further processing. In conjunction with other open source tools such as Deep Zoom and Caisis, this suite of tools is used for the management and archiving of digital microscopy images, enabling digitised images to be explored and zoomed online. Our online image repository also has the capacity to be used as a teaching resource. These tools also enable large files to be sectioned for image analysis.
Virtual slides: The virtual slide(s) for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/5330903258483934.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

