Stable nuclear transformation of Eudorina elegans
- PMID: 23402598
- PMCID: PMC3576287
- DOI: 10.1186/1472-6750-13-11
Stable nuclear transformation of Eudorina elegans
Abstract
Background: A fundamental step in evolution was the transition from unicellular to differentiated, multicellular organisms. Volvocine algae have been used for several decades as a model lineage to investigate the evolutionary aspects of multicellularity and cellular differentiation. There are two well-studied volvocine species, a unicellular alga (Chlamydomonas reinhardtii) and a multicellular alga with differentiated cell types (Volvox carteri). Species with intermediate characteristics also exist, which blur the boundaries between unicellularity and differentiated multicellularity. These species include the globular alga Eudorina elegans, which is composed of 16-32 cells. However, detailed molecular analyses of E. elegans require genetic manipulation. Unfortunately, genetic engineering has not yet been established for Eudorina, and only limited DNA and/or protein sequence information is available.
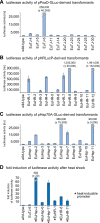
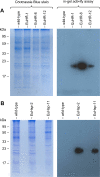
Results: Here, we describe the stable nuclear transformation of E. elegans by particle bombardment using both a chimeric selectable marker and reporter genes from different heterologous sources. Transgenic algae resistant to paromomycin were achieved using the aminoglycoside 3'-phosphotransferase VIII (aphVIII) gene of Streptomyces rimosus, an actinobacterium, under the control of an artificial promoter consisting of two V. carteri promoters in tandem. Transformants exhibited an increase in resistance to paromomycin by up to 333-fold. Co-transformation with non-selectable plasmids was achieved with a rate of 50 - 100%. The luciferase (gluc) gene from the marine copepod Gaussia princeps, which previously was engineered to match the codon usage of C. reinhardtii, was used as a reporter gene. The expression of gluc was mediated by promoters from C. reinhardtii and V. carteri. Heterologous heat shock promoters induced an increase in luciferase activity (up to 600-fold) at elevated temperatures. Long-term stability and both constitutive and inducible expression of the co-bombarded gluc gene was demonstrated by transcription analysis and bioluminescence assays.
Conclusions: Heterologous flanking sequences, including promoters, work in E. elegans and permit both constitutive and inducible expression of heterologous genes. Stable nuclear transformation of E. elegans is now routine. Thus, we show that genetic engineering of a species is possible even without the resources of endogenous genes and promoters.
Figures





Similar articles
-
Volvox and volvocine green algae.Evodevo. 2020 Jul 1;11:13. doi: 10.1186/s13227-020-00158-7. eCollection 2020. Evodevo. 2020. PMID: 32626570 Free PMC article. Review.
-
Stable nuclear transformation of Pandorina morum.BMC Biotechnol. 2014 Jul 17;14:65. doi: 10.1186/1472-6750-14-65. BMC Biotechnol. 2014. PMID: 25031031 Free PMC article.
-
Stable nuclear transformation of Gonium pectorale.BMC Biotechnol. 2009 Jul 10;9:64. doi: 10.1186/1472-6750-9-64. BMC Biotechnol. 2009. PMID: 19591675 Free PMC article.
-
A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii.Gene. 2001 Oct 17;277(1-2):221-9. doi: 10.1016/s0378-1119(01)00616-3. Gene. 2001. PMID: 11602359
-
The Genetics of Fitness Reorganization during the Transition to Multicellularity: The Volvocine regA-like Family as a Model.Genes (Basel). 2023 Apr 19;14(4):941. doi: 10.3390/genes14040941. Genes (Basel). 2023. PMID: 37107699 Free PMC article. Review.
Cited by
-
Transgene Expression in Microalgae-From Tools to Applications.Front Plant Sci. 2016 Apr 22;7:505. doi: 10.3389/fpls.2016.00505. eCollection 2016. Front Plant Sci. 2016. PMID: 27148328 Free PMC article. Review.
-
Volvox and volvocine green algae.Evodevo. 2020 Jul 1;11:13. doi: 10.1186/s13227-020-00158-7. eCollection 2020. Evodevo. 2020. PMID: 32626570 Free PMC article. Review.
-
Understanding nitrate assimilation and its regulation in microalgae.Front Plant Sci. 2015 Oct 26;6:899. doi: 10.3389/fpls.2015.00899. eCollection 2015. Front Plant Sci. 2015. PMID: 26579149 Free PMC article. Review.
-
Origins of multicellular complexity: Volvox and the volvocine algae.Mol Ecol. 2016 Mar;25(6):1213-23. doi: 10.1111/mec.13551. Epub 2016 Mar 1. Mol Ecol. 2016. PMID: 26822195 Free PMC article.
-
Development of a nuclear transformation system for Oleaginous Green Alga Lobosphaera (Parietochloris) incisa and genetic complementation of a mutant strain, deficient in arachidonic acid biosynthesis.PLoS One. 2014 Aug 18;9(8):e105223. doi: 10.1371/journal.pone.0105223. eCollection 2014. PLoS One. 2014. PMID: 25133787 Free PMC article.
References
-
- Grosberg RK, Strathmann R. The evolution of multicellularity: a minor major transition? Annu Rev Ecol Evol Syst. 2007;38:621–654. doi: 10.1146/annurev.ecolsys.36.102403.114735. - DOI
-
- Kirk DL. Volvox: molecular-genetic origins of multicellularity and cellular differentiation. 1. Cambridge University Press, Cambridge; 1998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

