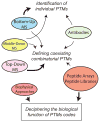
Deciphering post-translational modification codes
- PMID: 23402885
- PMCID: PMC3888991
- DOI: 10.1016/j.febslet.2013.01.047
Deciphering post-translational modification codes
Abstract
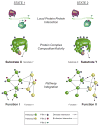
Post-translational modifications (PTMs) occur on nearly all proteins. Many domains within proteins are modified on multiple amino acid sidechains by diverse enzymes to create a myriad of possible protein species. How these combinations of PTMs lead to distinct biological outcomes is only beginning to be understood. This manuscript highlights several examples of combinatorial PTMs in proteins, and describes recent technological developments, which are driving our ability to understand how PTM patterns may "code" for biological outcomes.
Copyright © 2013 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. - PubMed
-
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, Shenoy SK, Gygi SP, Lefkowitz RJ. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Science signaling. 2011;4:ra51. - PMC - PubMed
-
- Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nature reviews Molecular cell biology. 2008;9:815–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

