A genome-wide RNA interference screen identifies new regulators of androgen receptor function in prostate cancer cells
- PMID: 23403032
- PMCID: PMC3613576
- DOI: 10.1101/gr.144774.112
A genome-wide RNA interference screen identifies new regulators of androgen receptor function in prostate cancer cells
Abstract
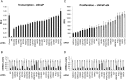
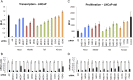
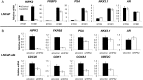
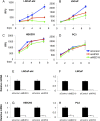
The androgen receptor (AR) is a mediator of both androgen-dependent and castration-resistant prostate cancers. Identification of cellular factors affecting AR transcriptional activity could in principle yield new targets that reduce AR activity and combat prostate cancer, yet a comprehensive analysis of the genes required for AR-dependent transcriptional activity has not been determined. Using an unbiased genetic approach that takes advantage of the evolutionary conservation of AR signaling, we have conducted a genome-wide RNAi screen in Drosophila cells for genes required for AR transcriptional activity and applied the results to human prostate cancer cells. We identified 45 AR-regulators, which include known pathway components and genes with functions not previously linked to AR regulation, such as HIPK2 (a protein kinase) and MED19 (a subunit of the Mediator complex). Depletion of HIPK2 and MED19 in human prostate cancer cells decreased AR target gene expression and, importantly, reduced the proliferation of androgen-dependent and castration-resistant prostate cancer cells. We also systematically analyzed additional Mediator subunits and uncovered a small subset of Mediator subunits that interpret AR signaling and affect AR-dependent transcription and prostate cancer cell proliferation. Importantly, targeting of HIPK2 by an FDA-approved kinase inhibitor phenocopied the effect of depletion by RNAi and reduced the growth of AR-positive, but not AR-negative, treatment-resistant prostate cancer cells. Thus, our screen has yielded new AR regulators including drugable targets that reduce the proliferation of castration-resistant prostate cancer cells.
Figures


References
-
- Al-Beiti MA, Lu X 2008. Expression of HIPK2 in cervical cancer: Correlation with clinicopathology and prognosis. Aust N Z J Obstet Gynaecol 48: 329–336 - PubMed
-
- Baidoobonso SM, Guidi BW, Myers LC 2007. Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae Mediator complex. J Biol Chem 282: 5551–5559 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
