Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor
- PMID: 23403292
- PMCID: PMC7116645
- DOI: 10.1016/j.celrep.2013.01.010
Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor
Abstract
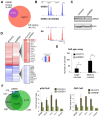
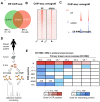
Estrogen receptor-α (ER) is the driving transcription factor in most breast cancers, and its associated proteins can influence drug response, but direct methods for identifying interacting proteins have been limited. We purified endogenous ER using an approach termed RIME (rapid immunoprecipitation mass spectrometry of endogenous proteins) and discovered the interactome under agonist- and antagonist-liganded conditions in breast cancer cells, revealing transcriptional networks in breast cancer. The most estrogen-enriched ER interactor is GREB1, a potential clinical biomarker with no known function. GREB1 is shown to be a chromatin-bound ER coactivator and is essential for ER-mediated transcription, because it stabilizes interactions between ER and additional cofactors. We show a GREB1-ER interaction in three xenograft tumors, and using a directed protein-protein approach, we find GREB1-ER interactions in half of ER(+) primary breast cancers. This finding is supported by histological expression of GREB1, which shows that GREB1 is expressed in half of ER(+) cancers, and predicts good clinical outcome. These findings reveal an unexpected role for GREB1 as an estrogen-specific ER cofactor that is expressed in drug-sensitive contexts.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
None of the authors have any conflicts of interest.
Figures


References
-
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nature reviews. 2002;2:101–112. - PubMed
-
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. Science. Vol. 277. New York, NY: 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer; pp. 965–968. - PubMed
-
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Dunbier AK, Anderson H, Ghazoui Z, Folkerd EJ, AHern R, Crowder RJ, Hoog J, Smith IE, Osin P, Nerurkar A, et al. Relationship between plasma estradiol levels and estrogen-responsive gene expression in estrogen receptor-positive breast cancer in postmenopausal women. J Clin Oncol. 2010;28:1161–1167. - PMC - PubMed
-
- Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer research. 2000;60:6367–6375. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

