Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance
- PMID: 23403935
- PMCID: PMC3565991
- DOI: 10.3389/fgene.2012.00260
Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance
Abstract
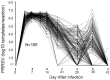
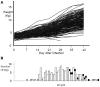
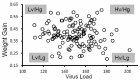
Infections caused by porcine reproductive and respiratory syndrome virus (PRRSV) have a severe economic impact on pig production in North America, Europe, and Asia. The emergence and eventual predominance of PRRS in the 1990s are the likely result of changes in the pork industry initiated in the late 1970s, which allowed the virus to occupy a unique niche within a modern commercial production system. PRRSV infection is responsible for severe clinical disease, but can maintain a life-long subclinical infection, as well as participate in several polymicrobial syndromes. Current vaccines lessen clinical signs, but are of limited use for disease control and elimination. The relatively poor protective immunity following vaccination is a function of the virus's capacity to generate a large degree of genetic diversity, combined with several strategies to evade innate and adaptive immune responses. In 2007, the PRRS Host Genetics consortium (PHGC) was established to explore the role of host genetics as an avenue for PRRS control. The PHGC model for PRRS incorporates the experimental infection of large numbers of growing pigs and has created the opportunity to study experimental PRRSV infection at the population level. The results show that pigs can be placed into distinct phenotypic groups, including pigs that show resistance (i.e., low virus load) or pigs that exhibit "tolerance" to infection. Tolerance was illustrated by pigs that gain weight normally in the face of a relatively high virus load. Genome-wide association analysis has identified a region on chromosome 4 (SSC4) correlated with resistance; i.e., lower cumulative virus load within the first 42 days of infection. The genomic region is near a family of genes involved in innate immunity. The region is also associated with higher weight gain in challenged pigs, suggesting that pigs with the resistance alleles don't seem to simultaneously experience reduction in growth, i.e., that resistance and tolerance are not antagonistically related. These results create the opportunity to develop breeding programs that will produce pigs with increased resistance to PRRS and simultaneously high growth rate. The identification of genomic markers involved in actual tolerance will likely prove more difficult, primarily because tolerance is difficult to quantify and because tolerance mechanism are still poorly understood. Another avenue of study includes the identification of genomic markers related to improved response following vaccination.
Keywords: PRRS resistance; genome-wide association study; porcine reproductive and respiratory syndrome.
Figures
References
-
- Benfield D. A., Nelson E., Collins J. E., Harris L., Goyal S. M., Robison D., et al. (1992). Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Invest. 4, 127–133 - PubMed
-
- Doeschl-Wilson A. B., Kyriazakis I., Vincent A., Rothschild M. F., Thacker E., Galina-Pantoja L. (2009). Clinical and pathological responses of pigs from two genetically diverse commercial lines to porcine reproductive and respiratory syndrome virus infection. J. Anim. Sci. 87, 1638–1647 10.2527/jas.2008-1447 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

