Structural constraints on the three-dimensional geometry of simple viruses: case studies of a new predictive tool
- PMID: 23403965
- PMCID: PMC3571114
- DOI: 10.1107/S0108767312047150
Structural constraints on the three-dimensional geometry of simple viruses: case studies of a new predictive tool
Abstract
Understanding the fundamental principles of virus architecture is one of the most important challenges in biology and medicine. Crick and Watson were the first to propose that viruses exhibit symmetry in the organization of their protein containers for reasons of genetic economy. Based on this, Caspar and Klug introduced quasi-equivalence theory to predict the relative locations of the coat proteins within these containers and classified virus structure in terms of T-numbers. Here it is shown that quasi-equivalence is part of a wider set of structural constraints on virus structure. These constraints can be formulated using an extension of the underlying symmetry group and this is demonstrated with a number of case studies. This new concept in virus biology provides for the first time predictive information on the structural constraints on coat protein and genome topography, and reveals a previously unrecognized structural interdependence of the shapes and sizes of different viral components. It opens up the possibility of distinguishing the structures of different viruses with the same T-number, suggesting a refined viral structure classification scheme. It can moreover be used as a basis for models of virus function, e.g. to characterize the start and end configurations of a structural transition important for infection.
Keywords: affine-extended symmetry group; genome organization; structural constraints; tiling theory; virus structure.
Figures
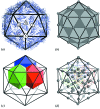
 surface lattice from quasi-equivalence theory that encodes the protein organizations in capsids composed of
surface lattice from quasi-equivalence theory that encodes the protein organizations in capsids composed of  coat proteins. (c), (d) A packing of overlapping icosahedra generates a partition of the icosahedral face akin to the
coat proteins. (c), (d) A packing of overlapping icosahedra generates a partition of the icosahedral face akin to the  structure: (c) shows three of the
structure: (c) shows three of the  translated and rotated icosahedra as solids; (d) shows the edges of these icosahedra, together with the vertices of all 60 icosahedra. This is an example of the structural constraints implied by our theory.
translated and rotated icosahedra as solids; (d) shows the edges of these icosahedra, together with the vertices of all 60 icosahedra. This is an example of the structural constraints implied by our theory.



References
-
- Bamford, D. H., Grimes, J. M. & Stuart, D. I. (2005). Curr. Opin. Struct. Biol. 15, 655–663. - PubMed
-
- Bruijn, N. G. de (1981a). Nederl. Akad. Wetensch. Indag. Math. 43, 39–52.
-
- Bruijn, N. G. de (1981b). Nederl. Akad. Wetensch. Indag. Math. 43, 53–66.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
