A decade of plague in Mahajanga, Madagascar: insights into the global maritime spread of pandemic plague
- PMID: 23404402
- PMCID: PMC3573667
- DOI: 10.1128/mBio.00623-12
A decade of plague in Mahajanga, Madagascar: insights into the global maritime spread of pandemic plague
Abstract
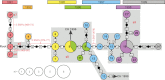
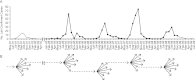
A cluster of human plague cases occurred in the seaport city of Mahajanga, Madagascar, from 1991 to 1999 following 62 years with no evidence of plague, which offered insights into plague pathogen dynamics in an urban environment. We analyzed a set of 44 Mahajanga isolates from this 9-year outbreak, as well as an additional 218 Malagasy isolates from the highland foci. We sequenced the genomes of four Mahajanga strains, performed whole-genome sequence single-nucleotide polymorphism (SNP) discovery on those strains, screened the discovered SNPs, and performed a high-resolution 43-locus multilocus variable-number tandem-repeat analysis of the isolate panel. Twenty-two new SNPs were identified and defined a new phylogenetic lineage among the Malagasy isolates. Phylogeographic analysis suggests that the Mahajanga lineage likely originated in the Ambositra district in the highlands, spread throughout the northern central highlands, and was then introduced into and became transiently established in Mahajanga. Although multiple transfers between the central highlands and Mahajanga occurred, there was a locally differentiating and dominant subpopulation that was primarily responsible for the 1991-to-1999 Mahajanga outbreaks. Phylotemporal analysis of this Mahajanga subpopulation revealed a cycling pattern of diversity generation and loss that occurred during and after each outbreak. This pattern is consistent with severe interseasonal genetic bottlenecks along with large seasonal population expansions. The ultimate extinction of plague pathogens in Mahajanga suggests that, in this environment, the plague pathogen niche is tenuous at best. However, the temporary large pathogen population expansion provides the means for plague pathogens to disperse and become ecologically established in more suitable nonurban environments.
Importance: Maritime spread of plague led to the global dissemination of this disease and affected the course of human history. Multiple historical plague waves resulted in massive human mortalities in three classical plague pandemics: Justinian (6th and 7th centuries), Middle Ages (14th to 17th centuries), and third (mid-1800s to the present). Key to these events was the pathogen's entry into new lands by "plague ships" via seaport cities. Although initial disease outbreaks in ports were common, they were almost never sustained for long and plague pathogens survived only if they could become established in ecologically suitable habitats. Although plague pathogens' ability to invade port cities has been essential for intercontinental spread, these regions have not proven to be a suitable long-term niche. The disease dynamics in port cities such as Mahajanga are thus critical to plague pathogen amplification and dispersal into new suitable ecological niches for the observed global long-term maintenance of plague pathogens.
Figures


References
-
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 101:17837–17842 - PMC - PubMed
-
- Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143 - PMC - PubMed
-
- Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, Zhang Y, Zheng H, Qin N, Xiao X, Wu M, Wang X, Zhou D, Qi Z, Du Z, Wu H, Yang X, Cao H, Wang H, Wang J, Yao S, Rakin A, Li Y, Falush D, Balloux F, Achtman M, Song Y, Wang J, Yang R. 2013. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 110:577–582 - PMC - PubMed
-
- Brygoo ER. 1966. Epidémiologie de la peste à Madagascar. Arch. Inst. Pasteur Madagascar 35:9–147
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

