Molecular characterization of basal-like and non-basal-like triple-negative breast cancer
- PMID: 23404817
- PMCID: PMC3579595
- DOI: 10.1634/theoncologist.2012-0397
Molecular characterization of basal-like and non-basal-like triple-negative breast cancer
Abstract
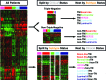
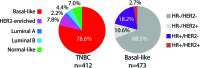
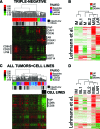
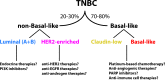
Triple-negative (TN) and basal-like (BL) breast cancer definitions have been used interchangeably to identify breast cancers that lack expression of the hormone receptors and overexpression and/or amplification of HER2. However, both classifications show substantial discordance rates when compared to each other. Here, we molecularly characterize TN tumors and BL tumors, comparing and contrasting the results in terms of common patterns and distinct patterns for each. In total, when testing 412 TN and 473 BL tumors, 21.4% and 31.5% were identified as non-BL and non-TN, respectively. TN tumors identified as luminal or HER2-enriched (HER2E) showed undistinguishable overall gene expression profiles when compared versus luminal or HER2E tumors that were not TN. Similar findings were observed within BL tumors regardless of their TN status, which suggests that molecular subtype is preserved regardless of individual marker results. Interestingly, most TN tumors identified as HER2E showed low HER2 expression and lacked HER2 amplification, despite the similar overall gene expression profiles to HER2E tumors that were clinically HER2-positive. Lastly, additional genomic classifications were examined within TN and BL cancers, most of which were highly concordant with tumor intrinsic subtype. These results suggest that future clinical trials focused on TN disease should consider stratifying patients based upon BL versus non-BL gene expression profiles, which appears to be the main biological difference seen in patients with TN breast cancer.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures




Comment in
-
Difference in therapeutic response between basal and nonbasal triple-negative breast cancers.Oncologist. 2013;18(9):1060-1. doi: 10.1634/theoncologist.2013-0121. Epub 2013 Aug 19. Oncologist. 2013. PMID: 23960187 Free PMC article.
References
-
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

